本文是Seurat包的学习笔记,相比以前的略有更新,重新整理。
2019年7月的生物信息学人才论坛会议上,伊现富老师说过一句话:“过去的流程使用的是过去的工具”,这次重新学单细胞,对这句话有了更深刻的理解。
是的,现在去看曾老板的视频还有豆豆之前的代码(2018-19年的),就有很多跑不通了。
并不是他们写错了,只是工具更新了,原来的函数和数据使用方法变了。
与时俱进这个词,在单细胞的领域体现的淋漓尽致,只有不断学习和进步,才能跟得上这变化的速度。
1.数据和R包准备
代码:https://satijalab.org/seurat/v3.0/pbmc3k_tutorial.html
数据:https://s3-us-west-2.amazonaws.com/10x.files/samples/cell/pbmc3k/pbmc3k_filtered_gene_bc_matrices.tar.gz
rm(list = ls())
options(stringsAsFactors = F)
library(dplyr)
library(Seurat)
library(patchwork)
2.读取数据
10X的输入数据是固定的三个文件,在工作目录下新建01_data/,把三个文件放进去。
dir("01_data/")
## [1] "barcodes.tsv" "genes.tsv" "matrix.mtx"
pbmc.data <- Read10X(data.dir = "01_data/")
pbmc <- CreateSeuratObject(counts = pbmc.data,
project = "pbmc3k",
min.cells = 3,
min.features = 200)
## Warning: Feature names cannot have underscores ('_'), replacing with dashes
## ('-')
单细胞相关的这几个R包都是打包成对象
pbmc
## An object of class Seurat
## 13714 features across 2700 samples within 1 assay
## Active assay: RNA (13714 features, 0 variable features)
pbmc.data[c("CD3D","TCL1A","MS4A1"), 1:30]
## 3 x 30 sparse Matrix of class "dgCMatrix"
## [[ suppressing 30 column names 'AAACATACAACCAC-1', 'AAACATTGAGCTAC-1', 'AAACATTGATCAGC-1' ... ]]
##
## CD3D 4 . 10 . . 1 2 3 1 . . 2 7 1 . . 1 3 . 2 3 . . . . . 3 4 1 5
## TCL1A . . . . . . . . 1 . . . . . . . . . . . . 1 . . . . . . . .
## MS4A1 . 6 . . . . . . 1 1 1 . . . . . . . . . 36 1 2 . . 2 . . . .
查看表达矩阵,很大一个。
exp = pbmc[["RNA"]]@counts;dim(exp)
## [1] 13714 2700
## [1] 13714 2700
exp[30:34,1:4]
## 5 x 4 sparse Matrix of class "dgCMatrix"
## AAACATACAACCAC-1 AAACATTGAGCTAC-1 AAACATTGATCAGC-1 AAACCGTGCTTCCG-1
## MRPL20 1 . 1 .
## ATAD3C . . . .
## ATAD3B . . . .
## ATAD3A . . . .
## SSU72 . 1 . 3
点号的地方是0,因为0太多了,存成稀疏矩阵可以省点空间。
3.质控
这里是对细胞进行的质控,指标是:
线粒体基因含量不能过高;
nFeature_RNA 不能过高或过低
为什么? nFeature_RNA是每个细胞中检测到的基因数量。nCount_RNA是细胞内检测到的分子总数。nFeature_RNA过低,表示该细胞可能已死/将死或是空液滴。太高的nCount_RNA和/或nFeature_RNA表明“细胞”实际上可以是两个或多个细胞。结合线粒体基因count数除去异常值,即可除去大多数双峰/死细胞/空液滴,因此它们过滤是常见的预处理步骤。 参考自:https://www.biostars.org/p/407036/
3.1 质控指标的可视化
pbmc[["percent.mt"]] <- PercentageFeatureSet(pbmc, pattern = "^MT-")
head(pbmc@meta.data, 3)
## orig.ident nCount_RNA nFeature_RNA percent.mt
## AAACATACAACCAC-1 pbmc3k 2419 779 3.0177759
## AAACATTGAGCTAC-1 pbmc3k 4903 1352 3.7935958
## AAACATTGATCAGC-1 pbmc3k 3147 1129 0.8897363
VlnPlot(pbmc,
features = c("nFeature_RNA",
"nCount_RNA",
"percent.mt"),
ncol = 3)
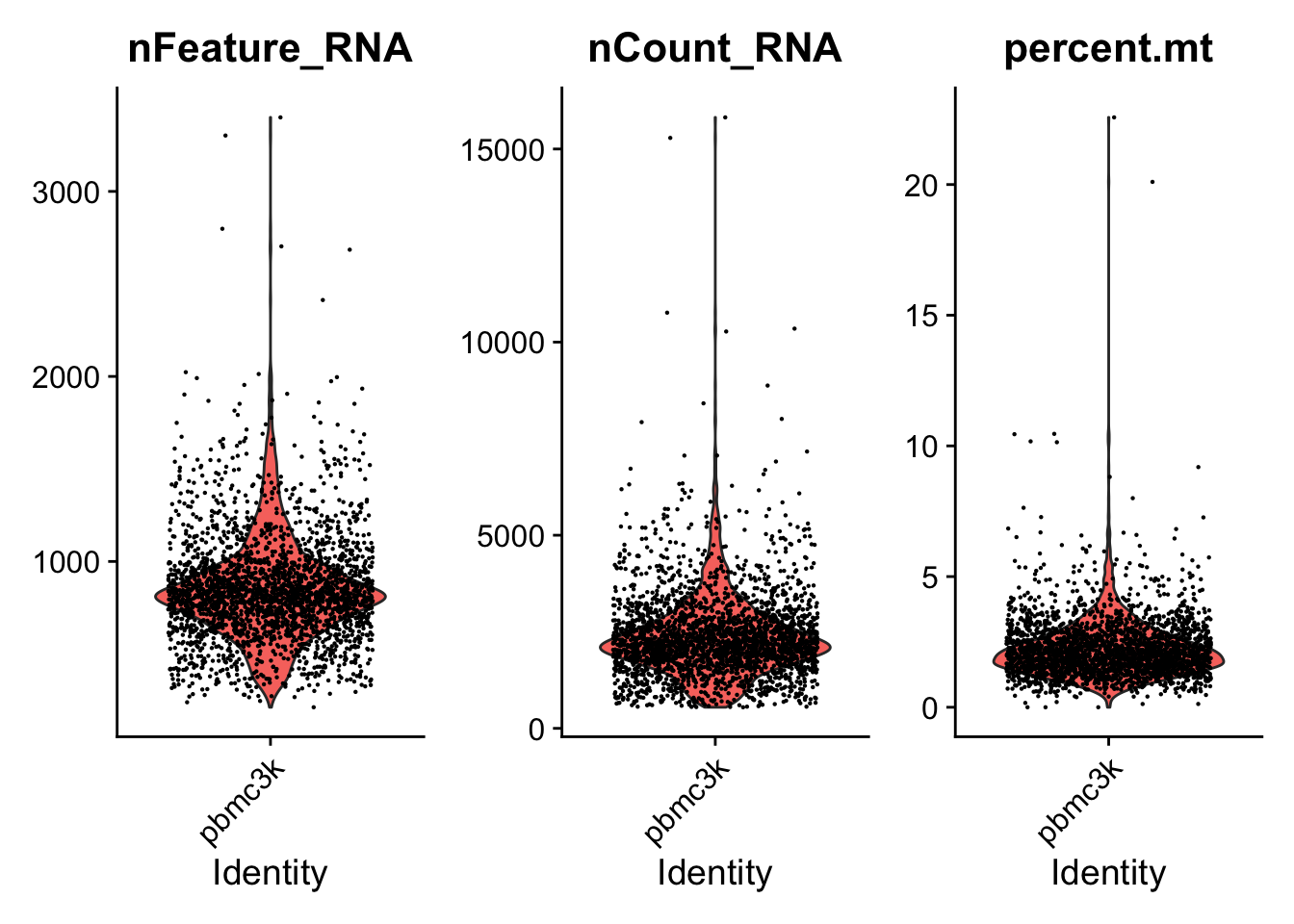
根据这个三个图,确定了这个数据的过滤标准:
nFeature_RNA在200~2500之间;线粒体基因占比在5%以下。
3.2 三个指标之间的相关性
plot1 <- FeatureScatter(pbmc,
feature1 = "nCount_RNA",
feature2 = "percent.mt")
plot2 <- FeatureScatter(pbmc,
feature1 = "nCount_RNA",
feature2 = "nFeature_RNA")
plot1 + plot2
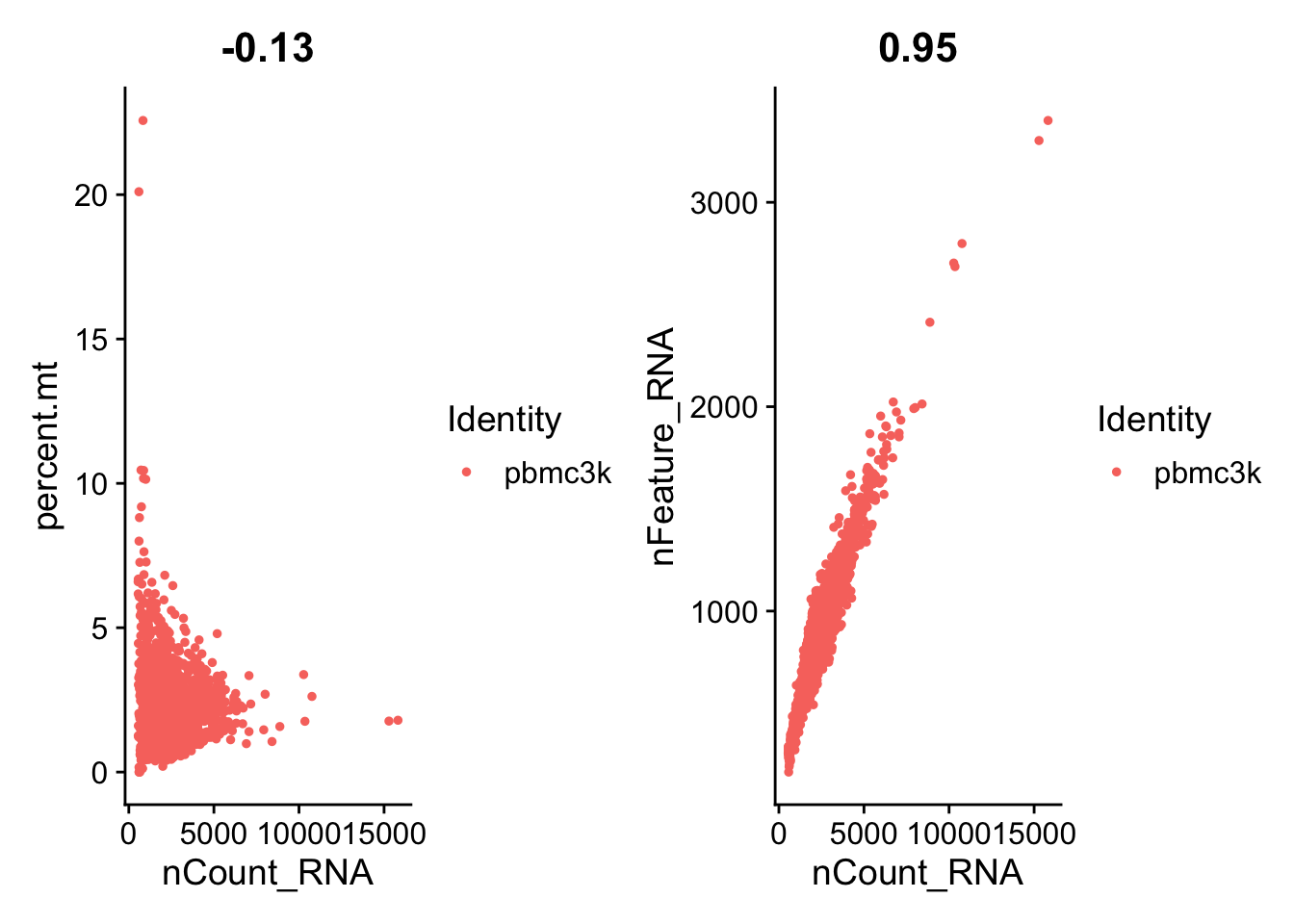
3.3 过滤
pbmc <- subset(pbmc,
subset = nFeature_RNA > 200 &
nFeature_RNA < 2500 &
percent.mt < 5)
dim(pbmc)
## [1] 13714 2638
原本是2700个细胞,可见用上面的标准是过滤掉了62个。
4. 数据归一化,找高变化基因
这里有点类似于常规转录组和芯片数据的差异分析过程,但并不是预知分组的比较,也没有具体的统计量来衡量,直接是你指定用哪种算法,要多少个高变化基因(HVG)
pbmc <- NormalizeData(pbmc)
pbmc@assays$RNA[30:34,1:3]
## 5 x 3 sparse Matrix of class "dgCMatrix"
## AAACATACAACCAC-1 AAACATTGAGCTAC-1 AAACATTGATCAGC-1
## MRPL20 1.635873 . 1.429744
## ATAD3C . . .
## ATAD3B . . .
## ATAD3A . . .
## SSU72 . 1.111715 .
# 有三种算法:vst、mean.var.plot、dispersion;默认选择2000个HVG
pbmc <- FindVariableFeatures(pbmc,
selection.method = "vst",
nfeatures = 2000)
top10 <- head(VariableFeatures(pbmc), 10);top10
## [1] "PPBP" "LYZ" "S100A9" "IGLL5" "GNLY" "FTL" "PF4" "FTH1"
## [9] "GNG11" "S100A8"
这里选了2000个,把前十个在图上标记出来。
plot1 <- VariableFeaturePlot(pbmc)
plot2 <- LabelPoints(plot = plot1,
points = top10,
repel = TRUE)
## When using repel, set xnudge and ynudge to 0 for optimal results
plot1 + plot2
## Warning: Transformation introduced infinite values in continuous x-axis
## Warning: Transformation introduced infinite values in continuous x-axis
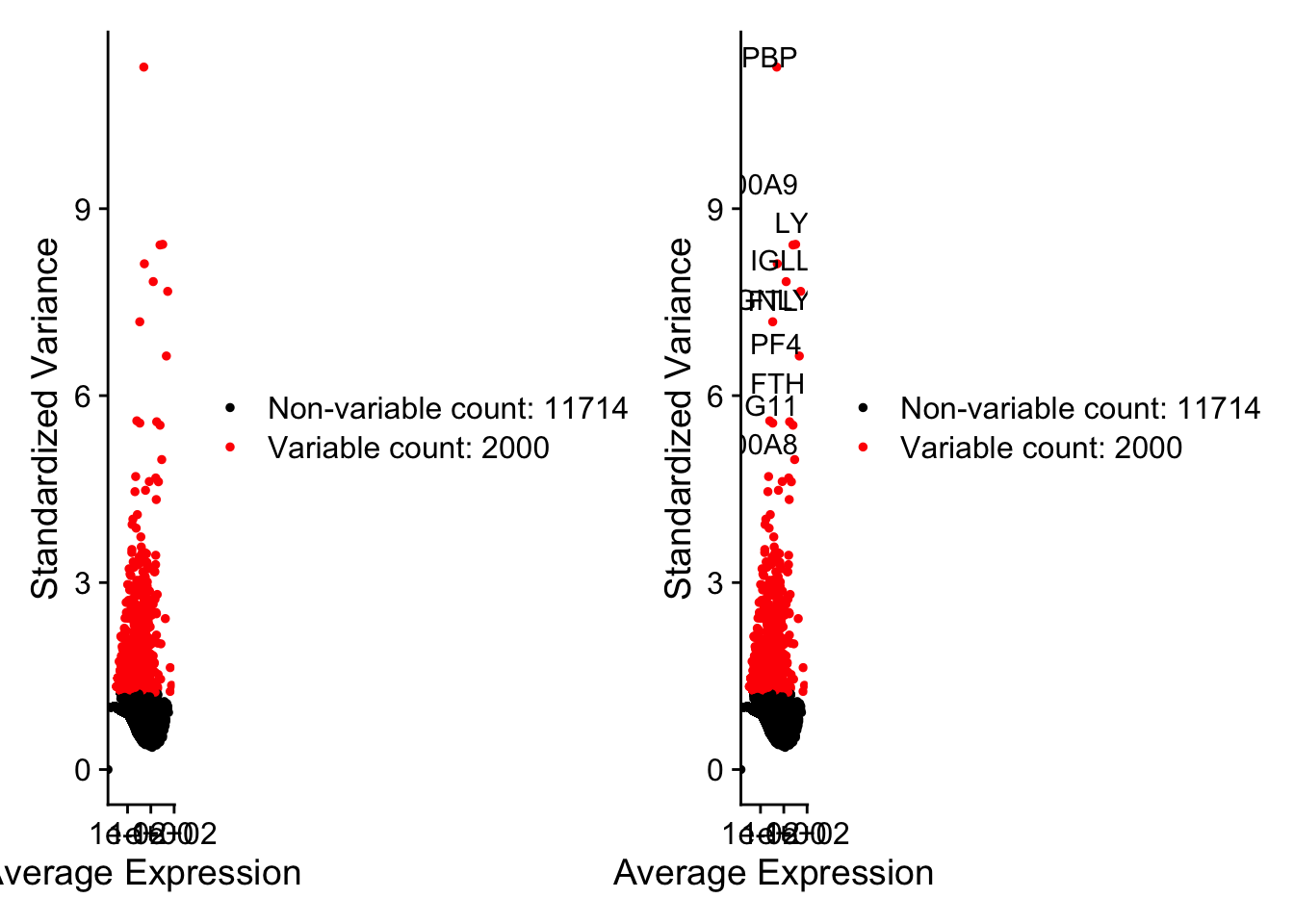
5. 标准化和降维
all.genes <- rownames(pbmc)
pbmc <- ScaleData(pbmc, features = all.genes)
## Centering and scaling data matrix
pbmc[["RNA"]]@scale.data[30:34,1:3]
曾经他们很多都是0。经过处理变成了不同的数字,这个并不会影响到后续分析,因为重点不是绝对的数字,而是比较起来的相对大小。为了让细胞与细胞之间的基因表达量更加可比,做这些处理是合理并可用的。
## AAACATACAACCAC-1 AAACATTGAGCTAC-1 AAACATTGATCAGC-1
## MRPL20 1.50566156 -0.56259931 1.24504892
## ATAD3C -0.04970561 -0.04970561 -0.04970561
## ATAD3B -0.10150202 -0.10150202 -0.10150202
## ATAD3A -0.13088200 -0.13088200 -0.13088200
## SSU72 -0.68454728 0.58087748 -0.68454728
ScaleData函数: 调整每个基因的表达量,使整个细胞的平均表达量为0 缩放每个基因的表达,从而使细胞之间的方差为1 此步骤给下游分析中的每个基因相等的权重,因此高表达的基因不会影响全局。
5.1 线性降维PCA
pbmc <- RunPCA(pbmc, features = VariableFeatures(object = pbmc))
## PC_ 1
## Positive: CST3, TYROBP, LST1, AIF1, FTL, FTH1, LYZ, FCN1, S100A9, TYMP
## FCER1G, CFD, LGALS1, S100A8, CTSS, LGALS2, SERPINA1, IFITM3, SPI1, CFP
## PSAP, IFI30, SAT1, COTL1, S100A11, NPC2, GRN, LGALS3, GSTP1, PYCARD
## Negative: MALAT1, LTB, IL32, IL7R, CD2, B2M, ACAP1, CD27, STK17A, CTSW
## CD247, GIMAP5, AQP3, CCL5, SELL, TRAF3IP3, GZMA, MAL, CST7, ITM2A
## MYC, GIMAP7, HOPX, BEX2, LDLRAP1, GZMK, ETS1, ZAP70, TNFAIP8, RIC3
## PC_ 2
## Positive: CD79A, MS4A1, TCL1A, HLA-DQA1, HLA-DQB1, HLA-DRA, LINC00926, CD79B, HLA-DRB1, CD74
## HLA-DMA, HLA-DPB1, HLA-DQA2, CD37, HLA-DRB5, HLA-DMB, HLA-DPA1, FCRLA, HVCN1, LTB
## BLNK, P2RX5, IGLL5, IRF8, SWAP70, ARHGAP24, FCGR2B, SMIM14, PPP1R14A, C16orf74
## Negative: NKG7, PRF1, CST7, GZMB, GZMA, FGFBP2, CTSW, GNLY, B2M, SPON2
## CCL4, GZMH, FCGR3A, CCL5, CD247, XCL2, CLIC3, AKR1C3, SRGN, HOPX
## TTC38, APMAP, CTSC, S100A4, IGFBP7, ANXA1, ID2, IL32, XCL1, RHOC
## PC_ 3
## Positive: HLA-DQA1, CD79A, CD79B, HLA-DQB1, HLA-DPB1, HLA-DPA1, CD74, MS4A1, HLA-DRB1, HLA-DRA
## HLA-DRB5, HLA-DQA2, TCL1A, LINC00926, HLA-DMB, HLA-DMA, CD37, HVCN1, FCRLA, IRF8
## PLAC8, BLNK, MALAT1, SMIM14, PLD4, LAT2, IGLL5, P2RX5, SWAP70, FCGR2B
## Negative: PPBP, PF4, SDPR, SPARC, GNG11, NRGN, GP9, RGS18, TUBB1, CLU
## HIST1H2AC, AP001189.4, ITGA2B, CD9, TMEM40, PTCRA, CA2, ACRBP, MMD, TREML1
## NGFRAP1, F13A1, SEPT5, RUFY1, TSC22D1, MPP1, CMTM5, RP11-367G6.3, MYL9, GP1BA
## PC_ 4
## Positive: HLA-DQA1, CD79B, CD79A, MS4A1, HLA-DQB1, CD74, HLA-DPB1, HIST1H2AC, PF4, TCL1A
## SDPR, HLA-DPA1, HLA-DRB1, HLA-DQA2, HLA-DRA, PPBP, LINC00926, GNG11, HLA-DRB5, SPARC
## GP9, AP001189.4, CA2, PTCRA, CD9, NRGN, RGS18, GZMB, CLU, TUBB1
## Negative: VIM, IL7R, S100A6, IL32, S100A8, S100A4, GIMAP7, S100A10, S100A9, MAL
## AQP3, CD2, CD14, FYB, LGALS2, GIMAP4, ANXA1, CD27, FCN1, RBP7
## LYZ, S100A11, GIMAP5, MS4A6A, S100A12, FOLR3, TRABD2A, AIF1, IL8, IFI6
## PC_ 5
## Positive: GZMB, NKG7, S100A8, FGFBP2, GNLY, CCL4, CST7, PRF1, GZMA, SPON2
## GZMH, S100A9, LGALS2, CCL3, CTSW, XCL2, CD14, CLIC3, S100A12, CCL5
## RBP7, MS4A6A, GSTP1, FOLR3, IGFBP7, TYROBP, TTC38, AKR1C3, XCL1, HOPX
## Negative: LTB, IL7R, CKB, VIM, MS4A7, AQP3, CYTIP, RP11-290F20.3, SIGLEC10, HMOX1
## PTGES3, LILRB2, MAL, CD27, HN1, CD2, GDI2, ANXA5, CORO1B, TUBA1B
## FAM110A, ATP1A1, TRADD, PPA1, CCDC109B, ABRACL, CTD-2006K23.1, WARS, VMO1, FYB
# 查看前三个主成分由哪些feature组成
print(pbmc[["pca"]], dims = 1:5, nfeatures = 5)
## PC_ 1
## Positive: CST3, TYROBP, LST1, AIF1, FTL
## Negative: MALAT1, LTB, IL32, IL7R, CD2
## PC_ 2
## Positive: CD79A, MS4A1, TCL1A, HLA-DQA1, HLA-DQB1
## Negative: NKG7, PRF1, CST7, GZMB, GZMA
## PC_ 3
## Positive: HLA-DQA1, CD79A, CD79B, HLA-DQB1, HLA-DPB1
## Negative: PPBP, PF4, SDPR, SPARC, GNG11
## PC_ 4
## Positive: HLA-DQA1, CD79B, CD79A, MS4A1, HLA-DQB1
## Negative: VIM, IL7R, S100A6, IL32, S100A8
## PC_ 5
## Positive: GZMB, NKG7, S100A8, FGFBP2, GNLY
## Negative: LTB, IL7R, CKB, VIM, MS4A7
VizDimLoadings(pbmc, dims = 1:2, reduction = "pca")
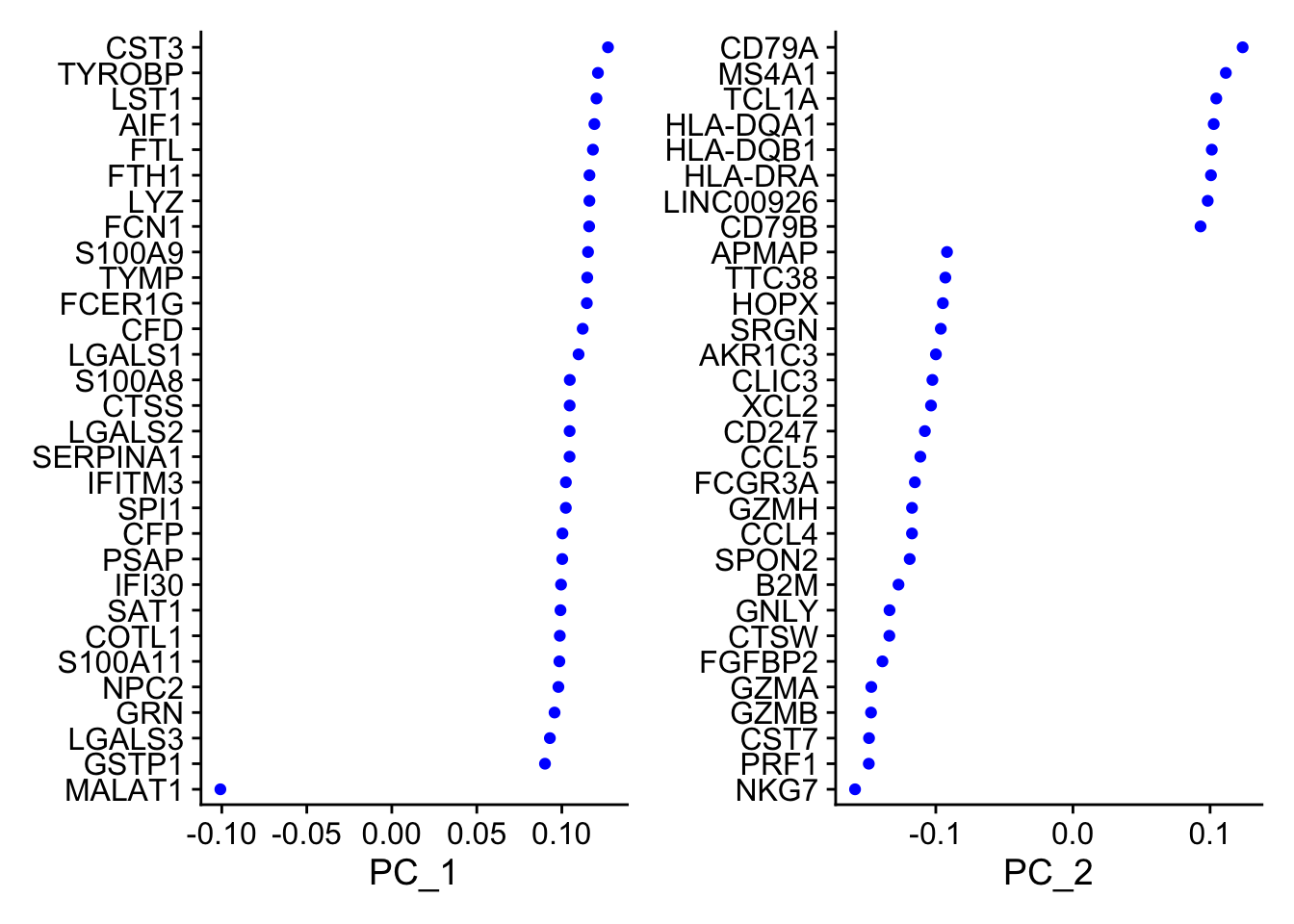
#每个主成分对应基因的热图
DimHeatmap(pbmc, dims = 1:15, cells = 500, balanced = TRUE)
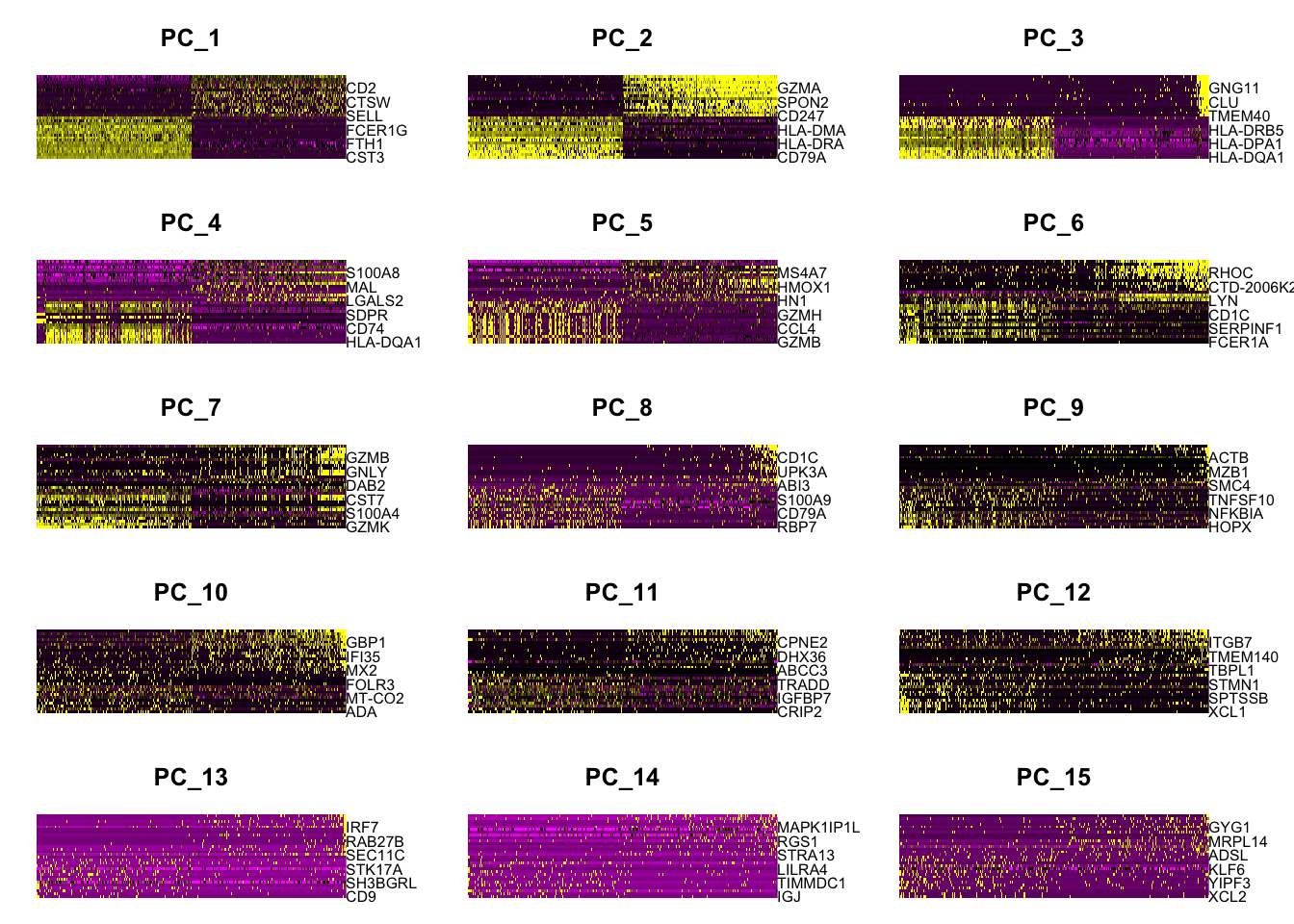
# 应该选多少个主成分进行后续分析
ElbowPlot(pbmc)
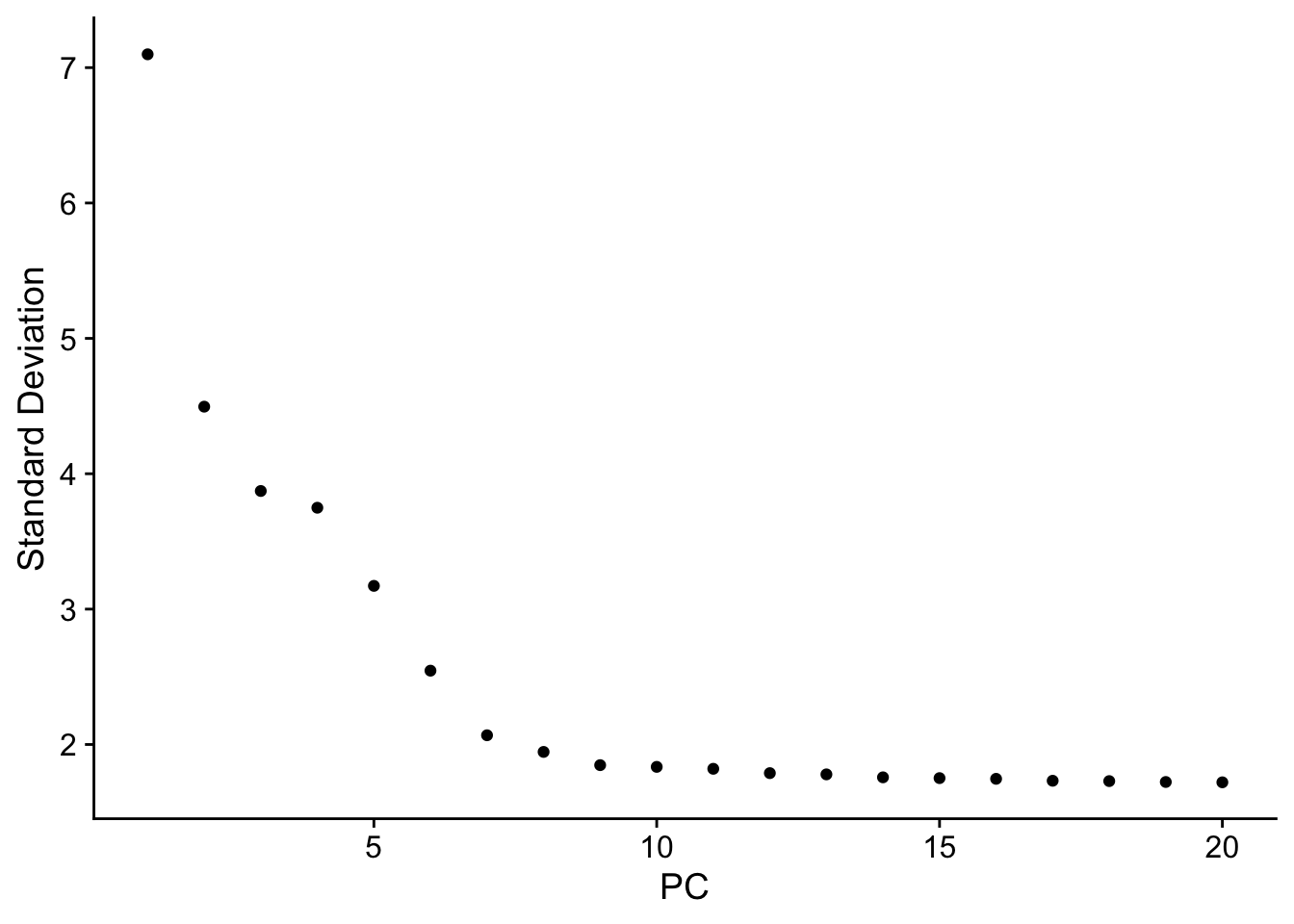
引用一下豆豆:它是根据每个主成分对总体变异水平的贡献百分比排序得到的图,我们主要关注”肘部“的PC,它是一个转折点(也即是这里的PC9-10),说明取前10个主成分可以比较好地代表总体变化
#PC1和2
DimPlot(pbmc, reduction = "pca")+ NoLegend()
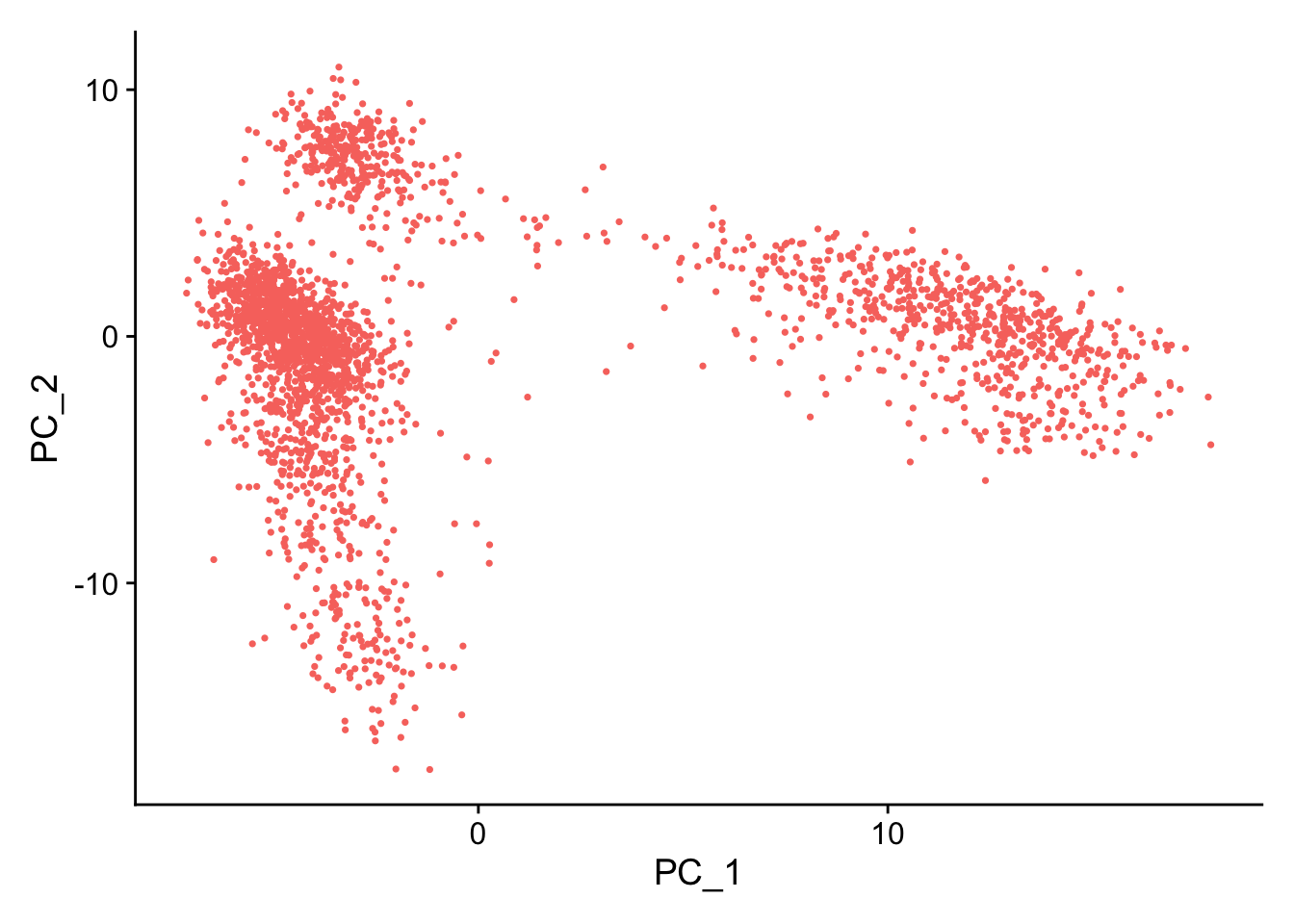
# 因为我们前面挑选了10个PCs,所以这里dims定义为10个
pbmc <- FindNeighbors(pbmc, dims = 1:10)
## Computing nearest neighbor graph
## Computing SNN
pbmc <- FindClusters(pbmc, resolution = 0.5) #分辨率
## Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
##
## Number of nodes: 2638
## Number of edges: 95965
##
## Running Louvain algorithm...
## Maximum modularity in 10 random starts: 0.8723
## Number of communities: 9
## Elapsed time: 0 seconds
# 结果聚成几类,用Idents查看
length(levels(Idents(pbmc)))
## [1] 9
resolution的大小决定着分群的数量,3000细胞时,0.4~1.2的分辨率表现较好。
5.2 UMAP 和t-sne
PCA是线性降维,这两个是非线性降维。理解起来比较复杂,用起来只是一个函数。
pbmc <- RunUMAP(pbmc, dims = 1:10)
## Warning: The default method for RunUMAP has changed from calling Python UMAP via reticulate to the R-native UWOT using the cosine metric
## To use Python UMAP via reticulate, set umap.method to 'umap-learn' and metric to 'correlation'
## This message will be shown once per session
## 14:28:12 UMAP embedding parameters a = 0.9922 b = 1.112
## 14:28:12 Read 2638 rows and found 10 numeric columns
## 14:28:12 Using Annoy for neighbor search, n_neighbors = 30
## 14:28:12 Building Annoy index with metric = cosine, n_trees = 50
## 0% 10 20 30 40 50 60 70 80 90 100%
## [----|----|----|----|----|----|----|----|----|----|
## **************************************************|
## 14:28:12 Writing NN index file to temp file /var/folders/mg/s3ln5v717ps4934qqym_3zk80000gn/T//RtmpWoykmm/file4458721f4d08
## 14:28:12 Searching Annoy index using 1 thread, search_k = 3000
## 14:28:13 Annoy recall = 100%
## 14:28:13 Commencing smooth kNN distance calibration using 1 thread
## 14:28:13 Initializing from normalized Laplacian + noise
## 14:28:13 Commencing optimization for 500 epochs, with 105124 positive edges
## 14:28:16 Optimization finished
DimPlot(pbmc, reduction = "umap")
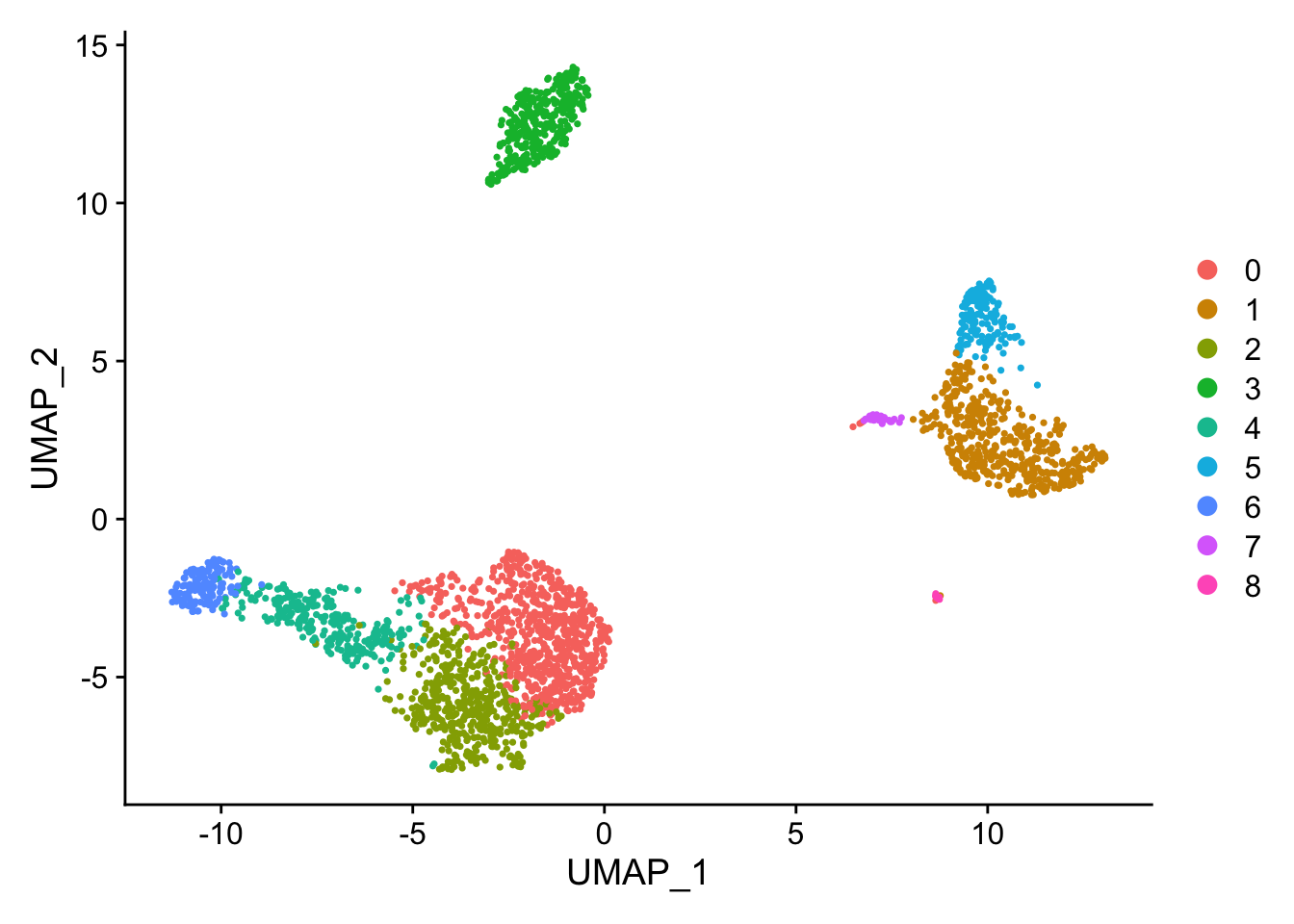
saveRDS(pbmc, file = "01_data/pbmc_tutorial.rds")
6.找marker基因
啥叫marker基因呢。和差异基因里面的上调基因有点类似,某个基因在某一簇细胞里表达量都很高,在其他簇表达量很低,那么这个基因就是这簇细胞的象征。
现成的函数可以实现:
- 单独找某个cluster的maker基因
- 比较某几个cluster
- 找全部cluster的maker基因
cluster1.markers <- FindMarkers(pbmc, ident.1 = 1, min.pct = 0.25)
head(cluster1.markers, n = 3)
## p_val avg_log2FC pct.1 pct.2 p_val_adj
## S100A9 0 5.570063 0.996 0.215 0
## S100A8 0 5.477394 0.975 0.121 0
## FCN1 0 3.394219 0.952 0.151 0
cluster5.markers <- FindMarkers(pbmc, ident.1 = 5, ident.2 = c(0, 3), min.pct = 0.25)
head(cluster5.markers, n = 3)
## p_val avg_log2FC pct.1 pct.2 p_val_adj
## FCGR3A 2.150929e-209 4.267579 0.975 0.039 2.949784e-205
## IFITM3 6.103366e-199 3.877105 0.975 0.048 8.370156e-195
## CFD 8.891428e-198 3.411039 0.938 0.037 1.219370e-193
pbmc.markers <- FindAllMarkers(pbmc, only.pos = TRUE, min.pct = 0.25, logfc.threshold = 0.25)
pbmc.markers %>% group_by(cluster) %>% top_n(n = 2, wt = avg_log2FC)
## Registered S3 method overwritten by 'cli':
## method from
## print.boxx spatstat.geom
## # A tibble: 18 x 7
## # Groups: cluster [9]
## p_val avg_log2FC pct.1 pct.2 p_val_adj cluster gene
## <dbl> <dbl> <dbl> <dbl> <dbl> <fct> <chr>
## 1 1.74e-109 1.07 0.897 0.593 2.39e-105 0 LDHB
## 2 1.17e- 83 1.33 0.435 0.108 1.60e- 79 0 CCR7
## 3 0. 5.57 0.996 0.215 0. 1 S100A9
## 4 0. 5.48 0.975 0.121 0. 1 S100A8
## 5 7.99e- 87 1.28 0.981 0.644 1.10e- 82 2 LTB
## 6 2.61e- 59 1.24 0.424 0.111 3.58e- 55 2 AQP3
## 7 0. 4.31 0.936 0.041 0. 3 CD79A
## 8 9.48e-271 3.59 0.622 0.022 1.30e-266 3 TCL1A
## 9 1.17e-178 2.97 0.957 0.241 1.60e-174 4 CCL5
## 10 4.93e-169 3.01 0.595 0.056 6.76e-165 4 GZMK
## 11 3.51e-184 3.31 0.975 0.134 4.82e-180 5 FCGR3A
## 12 2.03e-125 3.09 1 0.315 2.78e-121 5 LST1
## 13 1.05e-265 4.89 0.986 0.071 1.44e-261 6 GZMB
## 14 6.82e-175 4.92 0.958 0.135 9.36e-171 6 GNLY
## 15 1.48e-220 3.87 0.812 0.011 2.03e-216 7 FCER1A
## 16 1.67e- 21 2.87 1 0.513 2.28e- 17 7 HLA-DPB1
## 17 7.73e-200 7.24 1 0.01 1.06e-195 8 PF4
## 18 3.68e-110 8.58 1 0.024 5.05e-106 8 PPBP
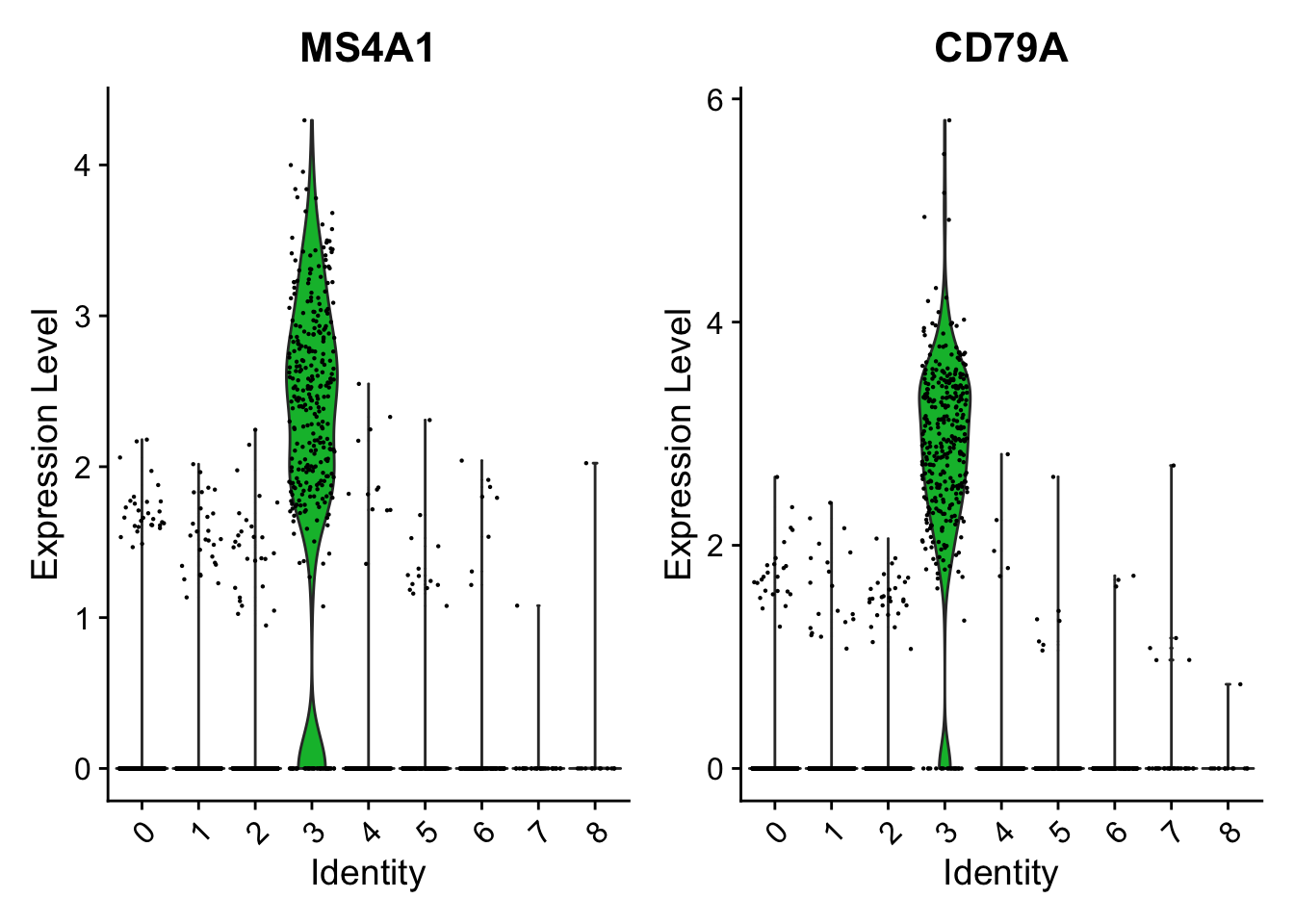
6.1 比较某个基因在几个cluster之间的表达量
小提琴图
VlnPlot(pbmc, features = c("MS4A1", "CD79A"))
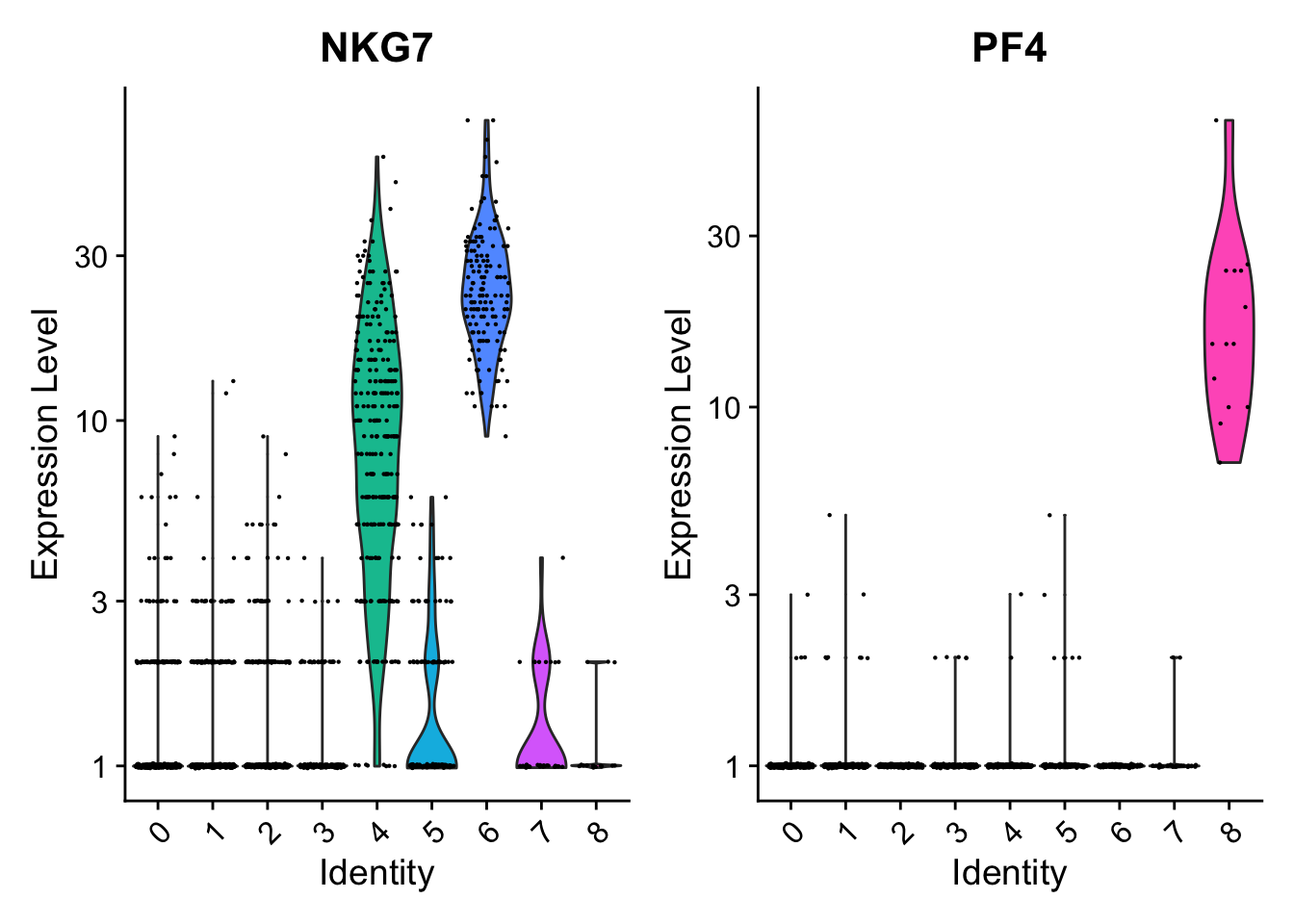
#可以拿count数据画
VlnPlot(pbmc, features = c("NKG7", "PF4"), slot = "counts", log = TRUE)
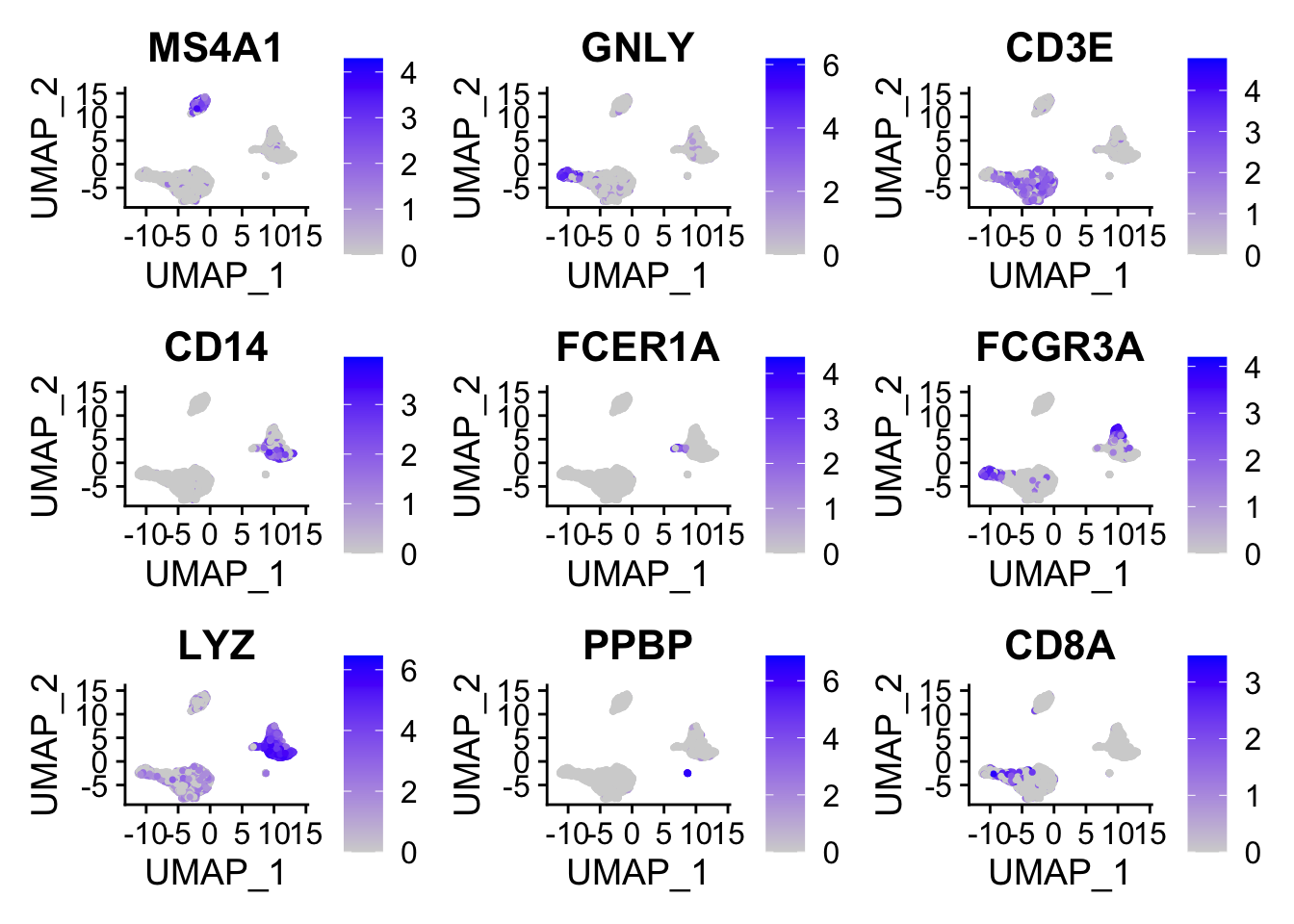
在umap图上标记
FeaturePlot(pbmc, features = c("MS4A1", "GNLY", "CD3E", "CD14", "FCER1A", "FCGR3A", "LYZ", "PPBP", "CD8A"))
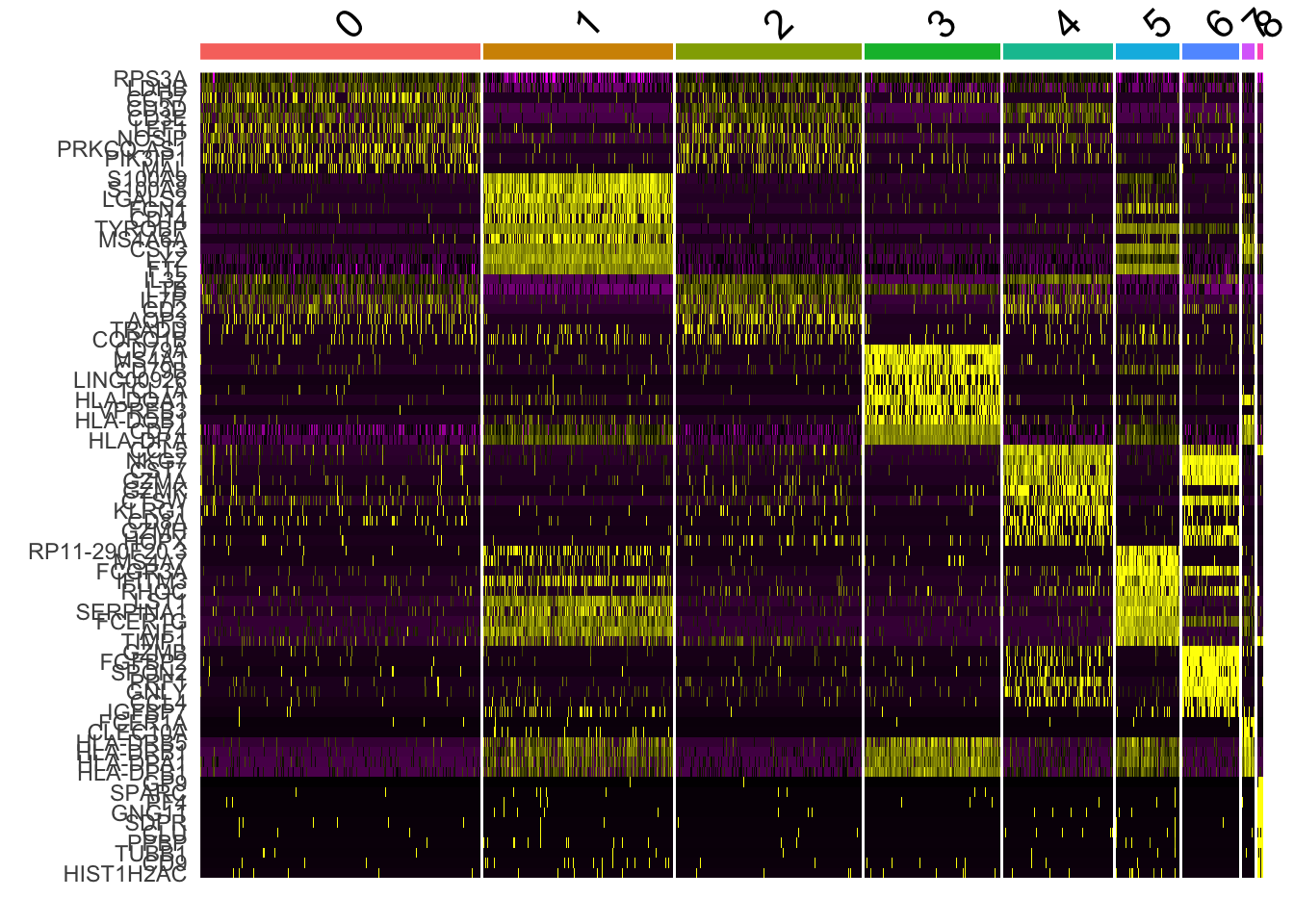
top10 <- pbmc.markers %>% group_by(cluster) %>% top_n(n = 10, wt = avg_log2FC)
6.2 marker基因的热图
DoHeatmap(pbmc, features = top10$gene) + NoLegend()
7. 根据marker基因确定细胞
在示例数据里,这一步找到哪些基因对应什么细胞的表格,是现成的。我问了豆豆,上哪里找这样的对应信息呢?豆豆说有专门的R包–singleR来做细胞注释,并且需要根据这些注释多次调整前面的参数,使聚类的结果与生物学意义对接上。标准操作会R语言就能搞定,生物学意义还需要慢慢磨啊。整理完三大R包,我就研究singleR。
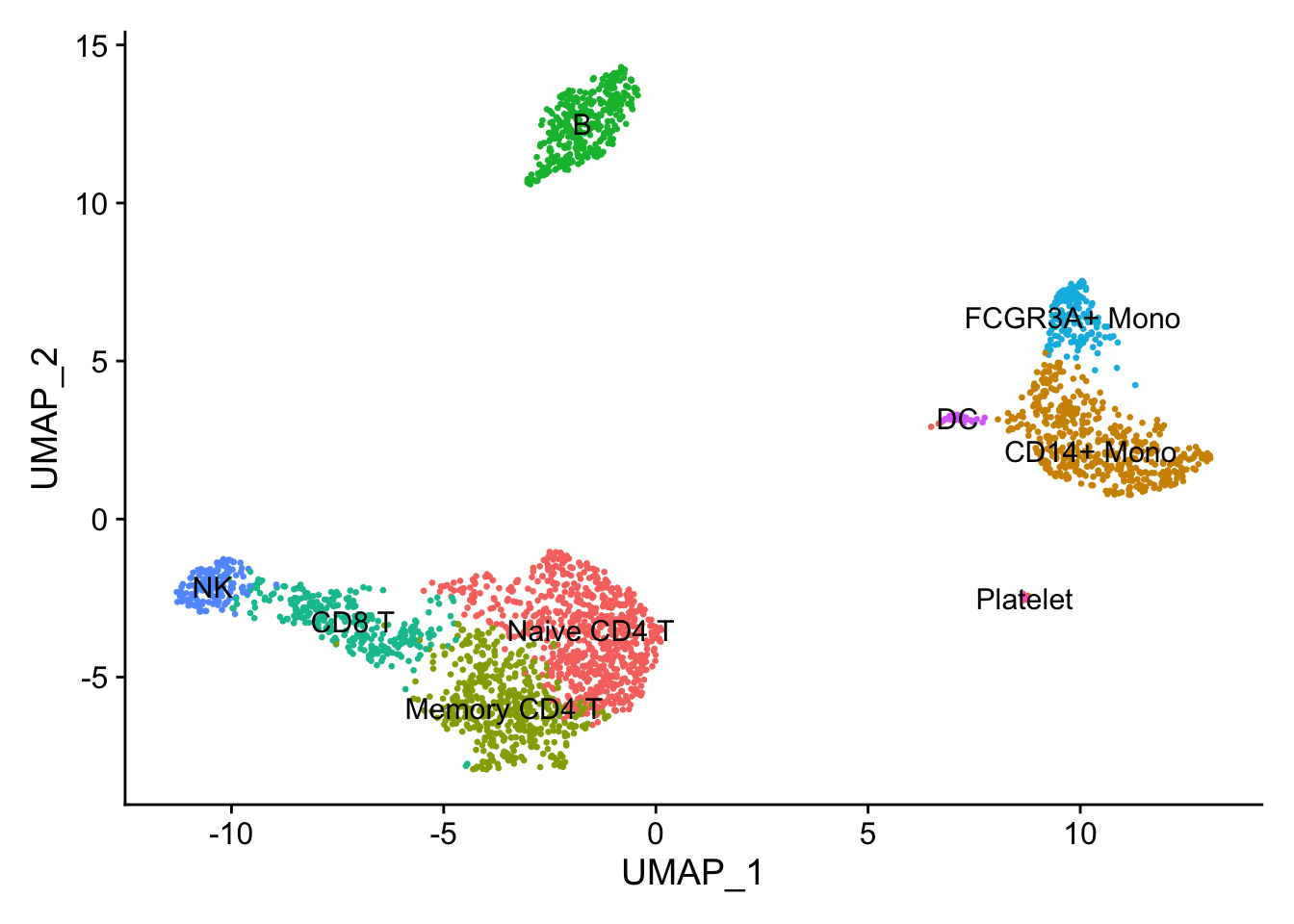
new.cluster.ids <- c("Naive CD4 T",
"CD14+ Mono",
"Memory CD4 T",
"B",
"CD8 T",
"FCGR3A+ Mono",
"NK",
"DC",
"Platelet")
names(new.cluster.ids) <- levels(pbmc)
pbmc <- RenameIdents(pbmc, new.cluster.ids)
DimPlot(pbmc,
reduction = "umap",
label = TRUE,
pt.size = 0.5) + NoLegend()
最后
以上就是安静鲜花最近收集整理的关于单细胞三大R包之Seurat的全部内容,更多相关单细胞三大R包之Seurat内容请搜索靠谱客的其他文章。








发表评论 取消回复