数据
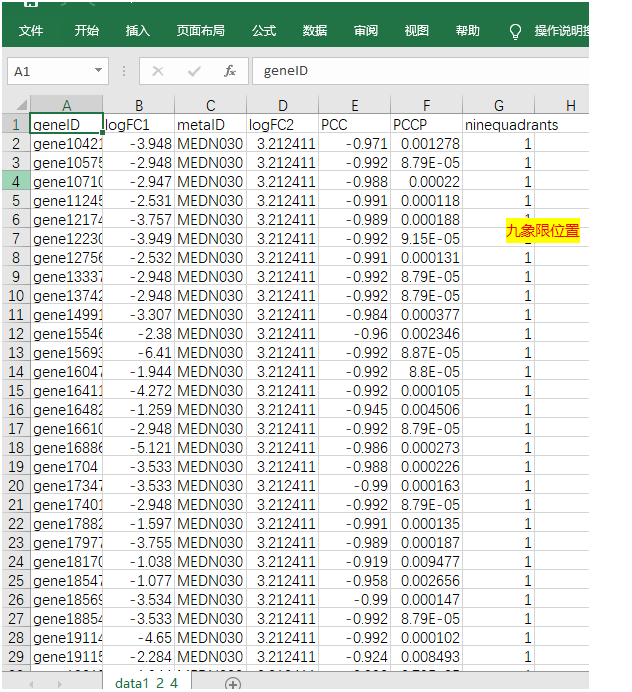
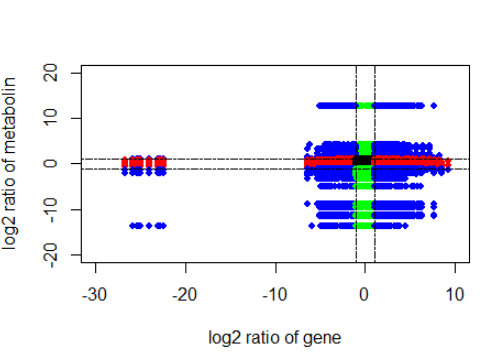
该数据包含基因与代谢物的logFC,基因与代谢物所持九象限的位置
第5象限没有,但是公司给的报告中显示黑色。下面是自己创建的第5象限数据,目的是把第5 象限涂黑
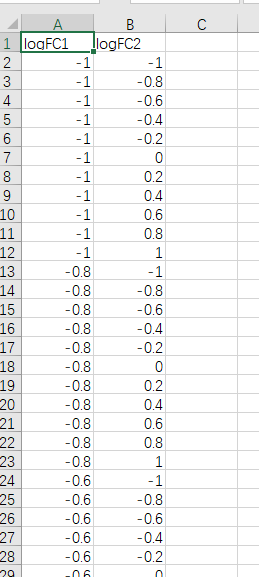
data1_2_4 <- read.csv("data1_2_4.csv",header = T)
data3_7 <- read.csv("data3_7.csv",header = T)
data6_8_9 <- read.csv("data6_8_9.csv",header = T)
data_1 <- data1_2_4[data1_2_4$ninequadrants==1,]
data_2 <- data1_2_4[data1_2_4$ninequadrants==2,]
data_4 <- data1_2_4[data1_2_4$ninequadrants==4,]
data_3 <- data3_7[data3_7$ninequadrants==3,]
data_7 <- data3_7[data3_7$ninequadrants==7,]
data_6 <- data6_8_9 [data6_8_9 $ninequadrants==6,]
data_8 <- data6_8_9 [data6_8_9 $ninequadrants==8,]
data_9 <- data6_8_9 [data6_8_9 $ninequadrants==9,]
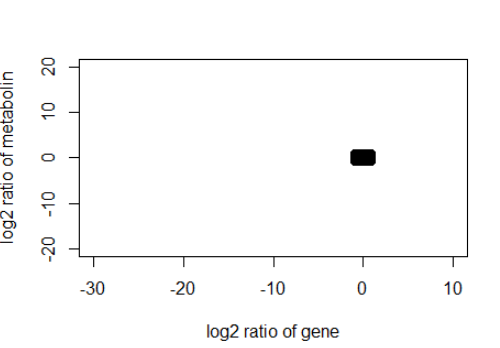
data_5 <- read.csv("data_5.csv",header = T)
plot(data_5$logFC1,data_5$logFC2,xlim = c(-30,10),ylim = c(-20,20),col = "black",pch=16,cex=0.8,xlab = "log2 ratio of gene",ylab = "log2 ratio of metabolin")
points(data_1$logFC1,data_1$logFC2,col="blue",pch=16,cex=0.8)
points(data_2$logFC1,data_2$logFC2,col="green",pch=16,cex=0.8)
points(data_3$logFC1,data_3$logFC2,col="blue",pch=16,cex=0.8)
points(data_4$logFC1,data_4$logFC2,col="red",pch=16,cex=0.8)
points(data_6$logFC1,data_6$logFC2,col="red",pch=16,cex=0.8)
points(data_7$logFC1,data_7$logFC2,col="blue",pch=16,cex=0.8)
points(data_8$logFC1,data_8$logFC2,col="green",pch=16,cex=0.8)
points(data_9$logFC1,data_9$logFC2,col="blue",pch=16,cex=0.8)
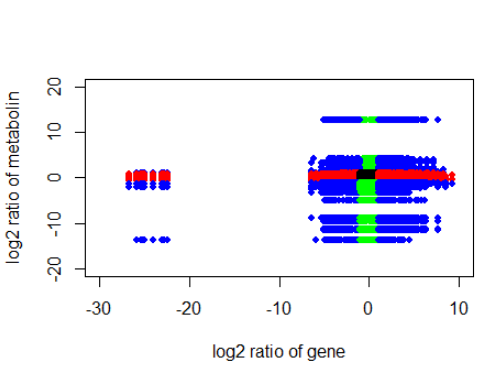
abline(v=c(-1,1),h=c(-1,1),lty=6)
最后
以上就是调皮咖啡豆最近收集整理的关于R绘制九象限图的全部内容,更多相关R绘制九象限图内容请搜索靠谱客的其他文章。
本图文内容来源于网友提供,作为学习参考使用,或来自网络收集整理,版权属于原作者所有。








发表评论 取消回复