python操作txt文件中数据教程[2]-python提取txt文件中的行列元素
觉得有用的话,欢迎一起讨论相互学习~




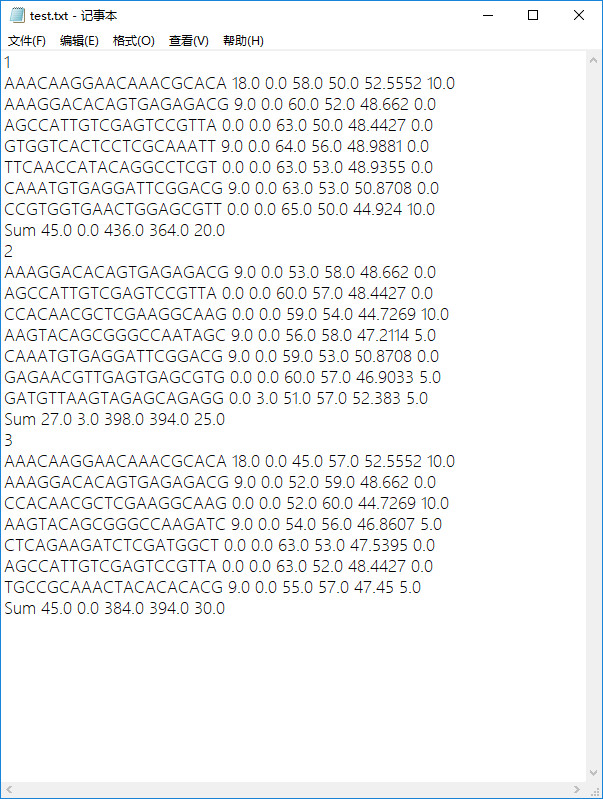
原始txt文件
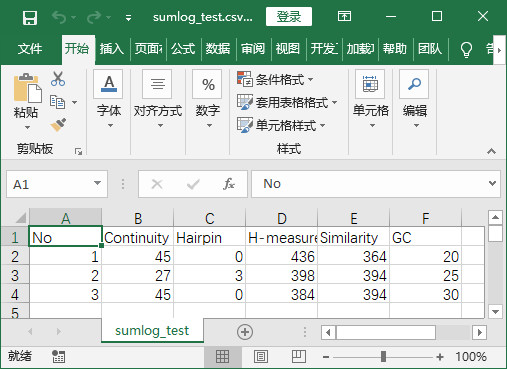
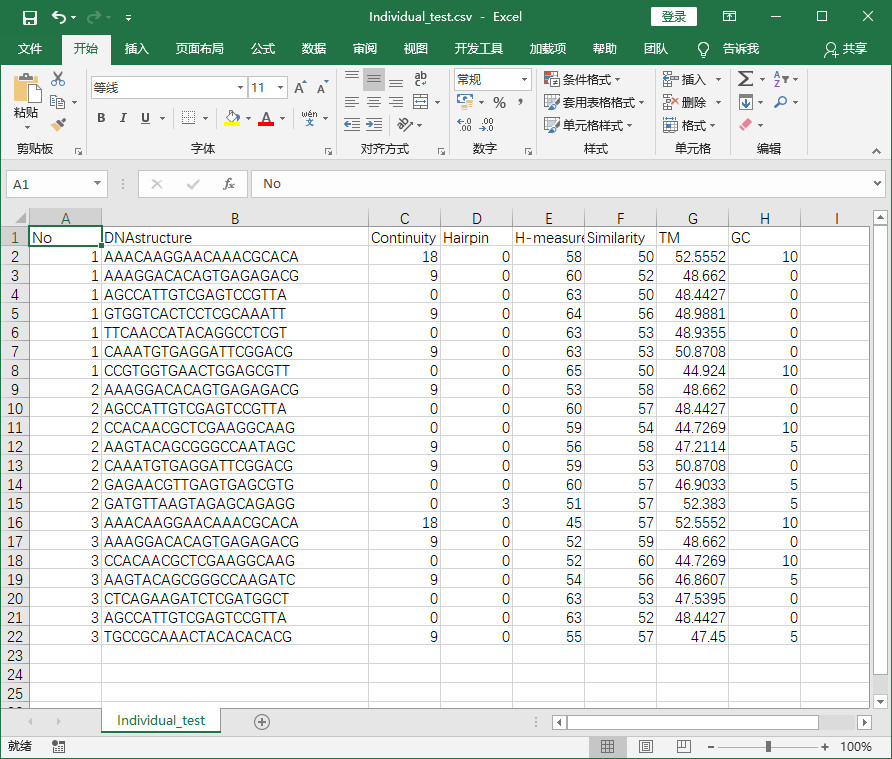
程序实现后结果-将txt中元素提取并保存在csv中
程序实现
import csv
filename = "./test/test.txt"
Sum_log_file = "./test/sumlog_test.csv"
Individual_log_file = "./test/Individual_test.csv"
DNA_log = [] # 精英种群个体日志mod9=1-8
Sum_log = [] # 精英种群总体日志mod9=0
DNA_Group = 7 # 表示每7条DNA组成一个组
# NO+'Sum 45.0 0.0 436.0 364.0 20.0n'中属性一共6个属性,,则设为8列的二维数组
sum_evaindex = [[] for i in range(6)]
# 个体有8个属性,则设为8列的二维数组
Individual_evaindex = [[] for i in range(8)]
# 将txt中文件信息保存到Sum_log和DNA_log列表中
with open(filename, 'r') as f:
i = 1
for line in f.readlines():
if i%9 == 0:
Sum_log.append(line)
else:
DNA_log.append(line)
i = i + 1
f.close()
# print(Sum_log)
# print(DNA_log)
# ['Sum 45.0 0.0 436.0 364.0 20.0n', 'Sum 27.0 3.0 398.0 394.0 25.0n', 'Sum 45.0 0.0 384.0 394.0 30.0']
# ['1n', 'AAACAAGGAACAAACGCACA 18.0 0.0 58.0 50.0 52.5552 10.0n', 'AAAGGACACAGTGAGAGACG 9.0 0.0 60.0 52.0 48.662 0.0n',
# 'AGCCATTGTCGAGTCCGTTA 0.0 0.0 63.0 50.0 48.4427 0.0n', 'GTGGTCACTCCTCGCAAATT 9.0 0.0 64.0 56.0 48.9881 0.0n',
# 'TTCAACCATACAGGCCTCGT 0.0 0.0 63.0 53.0 48.9355 0.0n', 'CAAATGTGAGGATTCGGACG 9.0 0.0 63.0 53.0 50.8708 0.0n',
# 'CCGTGGTGAACTGGAGCGTT 0.0 0.0 65.0 50.0 44.924 10.0n', '2n', 'AAAGGACACAGTGAGAGACG 9.0 0.0 53.0 58.0 48.662 0.0n',
# 'AGCCATTGTCGAGTCCGTTA 0.0 0.0 60.0 57.0 48.4427 0.0n', 'CCACAACGCTCGAAGGCAAG 0.0 0.0 59.0 54.0 44.7269 10.0n',
# 'AAGTACAGCGGGCCAATAGC 9.0 0.0 56.0 58.0 47.2114 5.0n', 'CAAATGTGAGGATTCGGACG 9.0 0.0 59.0 53.0 50.8708 0.0n',
# 'GAGAACGTTGAGTGAGCGTG 0.0 0.0 60.0 57.0 46.9033 5.0n', 'GATGTTAAGTAGAGCAGAGG 0.0 3.0 51.0 57.0 52.383 5.0n', '3n',
# 'AAACAAGGAACAAACGCACA 18.0 0.0 45.0 57.0 52.5552 10.0n', 'AAAGGACACAGTGAGAGACG 9.0 0.0 52.0 59.0 48.662 0.0n',
# 'CCACAACGCTCGAAGGCAAG 0.0 0.0 52.0 60.0 44.7269 10.0n', 'AAGTACAGCGGGCCAAGATC 9.0 0.0 54.0 56.0 46.8607 5.0n',
# 'CTCAGAAGATCTCGATGGCT 0.0 0.0 63.0 53.0 47.5395 0.0n', 'AGCCATTGTCGAGTCCGTTA 0.0 0.0 63.0 52.0 48.4427 0.0n',
# 'TGCCGCAAACTACACACACG 9.0 0.0 55.0 57.0 47.45 5.0n']
# 遍历行,并将列属性保存到对应列中
Sum_no = 1
for Sum in Sum_log:
# print(Sum.split("n")[0].split(" ")[1:])
# ['45.0', '0.0', '436.0', '364.0', '20.0']
# ['27.0', '3.0', '398.0', '394.0', '25.0']
# ['45.0', '0.0', '384.0', '394.0', '30.0']
sum_eva_index = Sum.split("n")[0].split(" ")[1:]
sum_evaindex[0].append(int(Sum_no))
sum_evaindex[1].append(float(sum_eva_index[0])) # Con
sum_evaindex[2].append(float(sum_eva_index[1])) # HP
sum_evaindex[3].append(float(sum_eva_index[2])) # Hm
sum_evaindex[4].append(float(sum_eva_index[3])) # Si
sum_evaindex[5].append(float(sum_eva_index[4])) # GC
Sum_no = Sum_no + 1
# print(sum_evaindex[0]) # [45.0, 27.0, 45.0]
# 遍历个体信息,并将其保存到Individual_evaindex列表中
dna_log_no = 0
for dna_log in DNA_log:
if (dna_log_no + 1)%8 == 1:
# print(int(dna_log.split("n")[0]))
# 以列存储序号值,并且重复DNA_Group次
for i in range(DNA_Group):
Individual_evaindex[0].append(int(dna_log.split("n")[0]))
else:
Individual_evaindex[1].append(dna_log.split("n")[0].split(" ")[0]) # 所有DNA序列全部记载,使用原有的str字符串类型记载
Individual_evaindex[2].append(float(dna_log.split("n")[0].split(" ")[1])) # DNA序列的连续值Con,注意要转换为浮点数类型
Individual_evaindex[3].append(float(dna_log.split("n")[0].split(" ")[2])) # Hp茎区匹配
Individual_evaindex[4].append(float(dna_log.split("n")[0].split(" ")[3])) # H-measure
Individual_evaindex[5].append(float(dna_log.split("n")[0].split(" ")[4])) # Similarity
Individual_evaindex[6].append(float(dna_log.split("n")[0].split(" ")[5])) # TM
Individual_evaindex[7].append(float(dna_log.split("n")[0].split(" ")[6])) # GC
dna_log_no = dna_log_no + 1
# print(Individual_evaindex[0]) #[1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3]
# print(Individual_evaindex[1])
# print(Individual_evaindex[2])
# print(Individual_evaindex[3])
# print(Individual_evaindex[4])
# print(Individual_evaindex[5])
# print(Individual_evaindex[6])
# print(Individual_evaindex[7])
# ['AAACAAGGAACAAACGCACA', 'AAAGGACACAGTGAGAGACG', 'AGCCATTGTCGAGTCCGTTA', 'GTGGTCACTCCTCGCAAATT', 'TTCAACCATACAGGCCTCGT',
# 'CAAATGTGAGGATTCGGACG', 'CCGTGGTGAACTGGAGCGTT', 'AAAGGACACAGTGAGAGACG', 'AGCCATTGTCGAGTCCGTTA', 'CCACAACGCTCGAAGGCAAG',
# 'AAGTACAGCGGGCCAATAGC', 'CAAATGTGAGGATTCGGACG', 'GAGAACGTTGAGTGAGCGTG', 'GATGTTAAGTAGAGCAGAGG', 'AAACAAGGAACAAACGCACA',
# 'AAAGGACACAGTGAGAGACG', 'CCACAACGCTCGAAGGCAAG', 'AAGTACAGCGGGCCAAGATC', 'CTCAGAAGATCTCGATGGCT', 'AGCCATTGTCGAGTCCGTTA',
# 'TGCCGCAAACTACACACACG']
# [18.0, 9.0, 0.0, 9.0, 0.0, 9.0, 0.0, 9.0, 0.0, 0.0, 9.0, 9.0, 0.0, 0.0, 18.0, 9.0, 0.0, 9.0, 0.0, 0.0, 9.0]
# [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 3.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
# [58.0, 60.0, 63.0, 64.0, 63.0, 63.0, 65.0, 53.0, 60.0, 59.0, 56.0, 59.0, 60.0, 51.0, 45.0, 52.0, 52.0, 54.0, 63.0, 63.0,
# 55.0]
# [50.0, 52.0, 50.0, 56.0, 53.0, 53.0, 50.0, 58.0, 57.0, 54.0, 58.0, 53.0, 57.0, 57.0, 57.0, 59.0, 60.0, 56.0, 53.0, 52.0,
# 57.0]
# [52.5552, 48.662, 48.4427, 48.9881, 48.9355, 50.8708, 44.924, 48.662, 48.4427, 44.7269, 47.2114, 50.8708, 46.9033,
# 52.383, 52.5552, 48.662, 44.7269, 46.8607, 47.5395, 48.4427, 47.45]
# [10.0, 0.0, 0.0, 0.0, 0.0, 0.0, 10.0, 0.0, 0.0, 10.0, 5.0, 0.0, 5.0, 5.0, 10.0, 0.0, 10.0, 5.0, 0.0, 0.0, 5.0]
Sum_log_file_header = ["No", "Continuity", "Hairpin", "H-measure", "Similarity", "GC"]
# 将数据写入csv日志文件中
with open(Sum_log_file, "w", newline='') as f:
writer = csv.writer(f)
writer.writerow(Sum_log_file_header) # 注意,此处使用writerow而不是使用writerows
for i in range(sum_evaindex[0][-1]): # i 取(0,1,2)
writer.writerow(
[sum_evaindex[0][i], sum_evaindex[1][i], sum_evaindex[2][i], sum_evaindex[3][i], sum_evaindex[4][i],
sum_evaindex[5][i]])
f.close()
Individual_log_file_header = ["No", "DNAstructure", "Continuity", "Hairpin", "H-measure", "Similarity", "TM", "GC"]
with open(Individual_log_file, "w", newline='') as f:
writer = csv.writer(f)
writer.writerow(Individual_log_file_header) # 注意,此处使用writerow而不是使用writerows
for i in range(sum_evaindex[0][-1]*DNA_Group):
writer.writerow(
[Individual_evaindex[0][i], Individual_evaindex[1][i], Individual_evaindex[2][i], Individual_evaindex[3][i],
Individual_evaindex[4][i], Individual_evaindex[5][i], Individual_evaindex[6][i],
Individual_evaindex[7][i]])
f.close()
测试版本
filename = "./test.txt"
DNA_log = [] # 精英种群个体日志mod9=2-8
Sum_log = [] # 精英种群总体日志mod9=0
Num_log = [] # 序号日志mod9=1
Num_int = [] # 截取序号为int类型
sum_evaindex = [[] for i in range(5)]
Individual_evaindex = [[] for i in range(8)]
with open(filename, 'r') as f:
i = 1
for line in f.readlines():
if i%9 == 1:
Num_log.append(line)
elif i%9 == 0:
Sum_log.append(line)
else:
DNA_log.append(line)
i = i + 1
f.close()
print(Num_log)
print(Num_log[1]) # 其中存着的不是数字1,而是字符串'2n',所以会有空行的情况
# ['1n', '2n', '3n']
# 2
#
#
print(Sum_log)
print(DNA_log)
# ['Sum 45.0 0.0 436.0 364.0 20.0n', 'Sum 27.0 3.0 398.0 394.0 25.0n', 'Sum 45.0 0.0 384.0 394.0 30.0']
# ['AAACAAGGAACAAACGCACA 18.0 0.0 58.0 50.0 52.5552 10.0n', 'AAAGGACACAGTGAGAGACG 9.0 0.0 60.0 52.0 48.662 0.0n',
# 'AGCCATTGTCGAGTCCGTTA 0.0 0.0 63.0 50.0 48.4427 0.0n', 'GTGGTCACTCCTCGCAAATT 9.0 0.0 64.0 56.0 48.9881 0.0n',
# 'TTCAACCATACAGGCCTCGT 0.0 0.0 63.0 53.0 48.9355 0.0n', 'CAAATGTGAGGATTCGGACG 9.0 0.0 63.0 53.0 50.8708 0.0n',
# 'CCGTGGTGAACTGGAGCGTT 0.0 0.0 65.0 50.0 44.924 10.0n', 'AAAGGACACAGTGAGAGACG 9.0 0.0 53.0 58.0 48.662 0.0n',
# 'AGCCATTGTCGAGTCCGTTA 0.0 0.0 60.0 57.0 48.4427 0.0n', 'CCACAACGCTCGAAGGCAAG 0.0 0.0 59.0 54.0 44.7269 10.0n',
# 'AAGTACAGCGGGCCAATAGC 9.0 0.0 56.0 58.0 47.2114 5.0n', 'CAAATGTGAGGATTCGGACG 9.0 0.0 59.0 53.0 50.8708 0.0n',
# 'GAGAACGTTGAGTGAGCGTG 0.0 0.0 60.0 57.0 46.9033 5.0n', 'GATGTTAAGTAGAGCAGAGG 0.0 3.0 51.0 57.0 52.383 5.0n',
# 'AAACAAGGAACAAACGCACA 18.0 0.0 45.0 57.0 52.5552 10.0n', 'AAAGGACACAGTGAGAGACG 9.0 0.0 52.0 59.0 48.662 0.0n',
# 'CCACAACGCTCGAAGGCAAG 0.0 0.0 52.0 60.0 44.7269 10.0n', 'AAGTACAGCGGGCCAAGATC 9.0 0.0 54.0 56.0 46.8607 5.0n',
# 'CTCAGAAGATCTCGATGGCT 0.0 0.0 63.0 53.0 47.5395 0.0n', 'AGCCATTGTCGAGTCCGTTA 0.0 0.0 63.0 52.0 48.4427 0.0n',
# 'TGCCGCAAACTACACACACG 9.0 0.0 55.0 57.0 47.45 5.0n']
for no in Num_log:
# print(no[0]) # 字符形式的数字1,这是错的,因为有可能序号超过一位数
# Num_int.append(int(no.split("n"))) ['1', '']
Num_int.append(int(no.split("n")[0]))
for Sum in Sum_log:
# print(Sum.split("n")[0].split(" ")[1:])
# ['45.0', '0.0', '436.0', '364.0', '20.0']
# ['27.0', '3.0', '398.0', '394.0', '25.0']
# ['45.0', '0.0', '384.0', '394.0', '30.0']
sum_eva_index = Sum.split("n")[0].split(" ")[1:]
sum_evaindex[0].append(float(sum_eva_index[0]))
sum_evaindex[1].append(float(sum_eva_index[1]))
sum_evaindex[2].append(float(sum_eva_index[2]))
sum_evaindex[3].append(float(sum_eva_index[3]))
sum_evaindex[4].append(float(sum_eva_index[4]))
print(sum_evaindex[0]) # [45.0, 27.0, 45.0]
最后
以上就是温柔西装最近收集整理的关于python提取txt中文数据_python操作txt文件中数据教程[2]-python提取txt文件的全部内容,更多相关python提取txt中文数据_python操作txt文件中数据教程[2]-python提取txt文件内容请搜索靠谱客的其他文章。




![python提取txt中文数据_python操作txt文件中数据教程[2]-python提取txt文件](https://file2.kaopuke.com:8081/files_image/reation/bcimg3.png)
![mysql sqlcode 2014_SQLSTATE[HY000]: General error: 2014 Cannot execute queries while other unbuffere...](https://file2.kaopuke.com:8081/files_image/reation/bcimg4.png)


发表评论 取消回复