关于微阵列芯片和RNA-seq的比较
转录组代表存在于细胞中RNA的全部类型,包括mRNA、rRNA、tRNA以及其它各种非编码RNA等。转录组是了解细胞过程的主要手段,微阵列(Microarray)和RNA-seq(RNA sequencing)是转录组分析中的两种主要技术。它们的主要区别在于,微阵列基于预先设计的标记探针与目标cDNA序列的杂交,而RNA-seq通过测序技术对cDNA链进行直接测序。
微阵列
微阵列取决于杂交探针,根据杂交信号强度定量基因相对表达水平。
首先从样品中提取总RNA,然后构建cDNA文库。将cDNA与预先设计的带荧光标记的DNA探针在固体表面(spot matrix)混合,互补序列将与微阵列中的标记探针杂交。通过检测微阵列的探针荧光强度,这些探针代表了不同的基因,可获得各基因的相对表达谱。
一般而言,微阵列探针的荧光强度应与样品中互补cDNA(代表转录物)的丰度成正比。但是该技术的准确性取决于所设计的探针,已知序列的碱基组成以及探针杂交的亲和力,因此具有局限性。微阵列技术不能用低丰度的转录本进行,且不能区分同工型以及鉴定遗传变异。此外,探针也常伴随交叉杂交,非特异性杂交等问题。
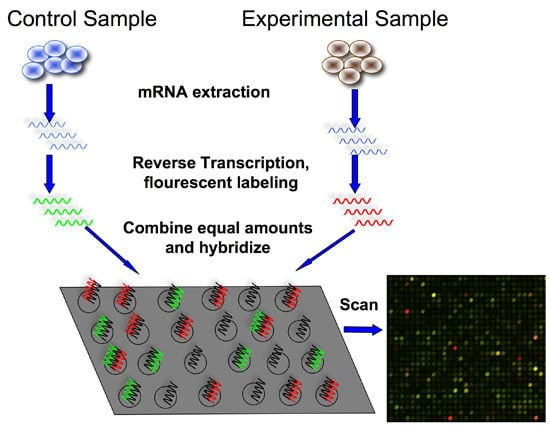
微阵列芯片
RNA-seq
RNA-seq是一种快速且高通量的方法,不依赖于预先设计的探针或已知的序列碱基特征,因此具有较高的灵敏度和检测新基因以及遗传变异的能力。
首先提取总RNA并在纯化后片段化处理,然后构建cDNA文库,使用高通量测序的方法对cDNA进行测序。获得测序序列后比对到参考基因组上(或者从头组装转录本后再比对),根据测序序列的覆盖情况定量基因表达。
理论上,如果测序深度足够深,则可以覆盖到所有基因,包括尚未发现的新基因,并能实现全长基因水平的定量。并且RNA-seq本身是一种测序技术,能获得基因的碱基组成信息,因此除了用于定量基因表达外,还可用于基因组结构研究,这是微阵列芯片无法比拟的。但是高通量的方法存在一定的假阳性问题是不可忽视的,并且二代测序在建库时也存在非特异性扩增的偏移,三代测序定量相对准确但价格昂贵。
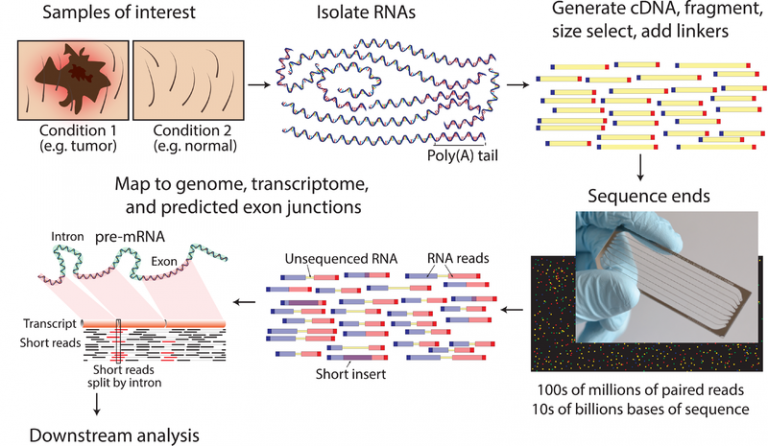
RNA测序(RNA-seq)
微阵列和RNA-seq的比较
就目前来说,小编比较推荐使用RNA-seq进行转录谱研究。一是数据量大,覆盖基因组范围更广;二是不受物种基因组是否已知所限制;三是RNA-seq数据的灵活度高,并能用于基因组结构分析。

微阵列和RNA-seq的比较
参考链接
https://www.differencebetween.com/difference-between-microarray-and-vs-rna-sequencing/
Difference Between Microarray and RNA Sequencing
March 17, 2017 Posted by Samanthi
3
Key Difference – Microarray vs RNA Sequencing
Transcriptome represents the whole content of RNA present in a cell including mRNA, rRNA, tRNA, degraded RNA, and, nondegraded RNA. Profiling transcriptome is an important process in order to understand the cell insights. There are several advanced methods for transcriptome profiling. Microarray and RNA sequencing are two types of technologies developed to analyze transcriptome. The key difference between microarray and RNA sequencing is that microarray is based on the hybridization potential of predesigned labeled probes with target cDNA sequences while RNA sequencing is based on the direct sequencing of cDNA strands by advanced sequencing techniques such as NGS. Microarray is performed with the prior knowledge about the sequences and RNA sequencing is performed without the prior knowledge about sequences.
CONTENTS
1. Overview and Key Difference
2. What is Microarray
3. What is RNA Sequencing
4. Side by Side Comparison – Microarray vs RNA Sequencing
5. Summary
What is Microarray?
Microarray is a robust, reliable and high throughput method used for transcriptome profiling by scientists. It is the most popular approach for transcript analysis. It is a low-cost method, which depends on the hybridization probes.
The technique starts with extraction of mRNA from the sample and the construction of cDNA library from total RNA. Then it is mixed with fluorescently labeled predesigned probes on a solid surface (spot matrix). Complementary sequences hybridize with the labeled probes in the microarray. Then microarray is washed and screened, and the image is quantified. Gathered data should be analyzed to get the relative expression profiles.
The intensity of the microarray probes is assumed to be proportional to the quantity of transcripts in the sample. However, the accuracy of the technique depends on the designed probes, prior knowledge of the sequence and the affinity of probes for hybridization. Hence microarray technology has limitations. Microarray technique cannot be performed with low abundance transcripts. It fails to differentiate isoforms and identify genetic variants. Since this method depends on hybridization of probes, some problems related to hybridization such as cross-hybridization, nonspecific hybridization etc. occurs in microarray technique.
Figure 01: Microarray
What is RNA Sequencing?
RNA shotgun sequencing (RNA seq) is a recently developed whole transcriptome sequencing technique. It’s a rapid and high throughput method of transcriptome profiling. It directly quantifies the expression of genes and results in deep investigation of the transcriptome. RNA seq does not depend on predesigned probes or prior knowledge of the sequences. Therefore, RNA seq method has high sensitivity and capability of detecting novel genes and genetic variants.
RNA sequencing method is performed via several steps. Total RNA of the cell must be isolated and fragmented. Then, using reverse transcriptase, a cDNA library must be prepared. Each cDNA strand must be ligated with adaptors. Then the ligated fragments must be amplified and purified. Finally using a NGS method, sequencing of the cDNA must be performed.
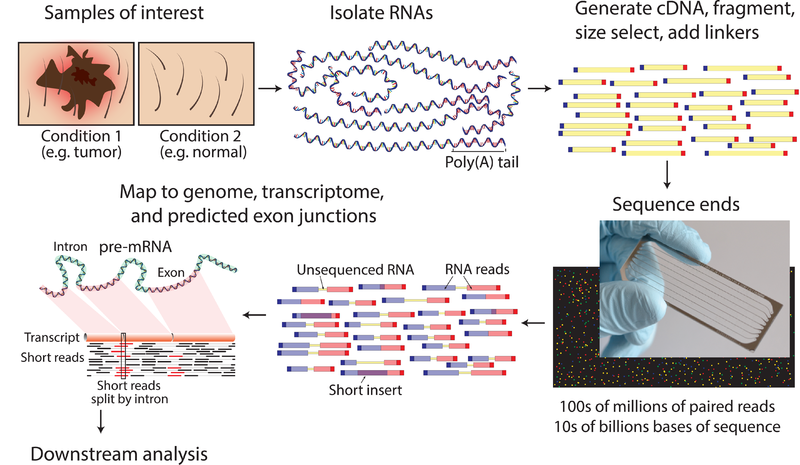
Figure 02: RNA Sequencing
What is the difference between Microarray and RNA sequencing?
Microarray vs RNA Sequencing | |
| Microarray is a robust, reliable, high throughput method. | RNA sequencing is an accurate and high-throughput method. |
| Cost | |
| This is a low-cost method. | This is an expensive method. |
| Analysis of a Large Number of Samples | |
| This facilitates analyzing a large number of samples simultaneously. | This facilitates analyzing a large number of samples. |
| Data Analysis | |
| Data analysis is complex. | More data is generated in this method; hence, the process is more complex. |
| Prior Knowledge of Sequences | |
| This method is based on hybridization probes, so prior knowledge of sequences in required. | This method does not depend on the prior sequence knowledge. |
| Structural Variations and Novel Genes | |
| This method cannot detect structural variations and novel genes. | This method can detect structural variations such as gene fusing, alternative splicing, and novel genes. |
| Sensitivity | |
| This cannot detect differences in expression of isoforms, so this has limited sensitivity. | This has high sensitivity. |
| Outcome | |
| This can only result in relative expression levels. This does not give absolute quantification of gene expression. | It gives absolute and relative expression levels. |
| Data Reanalysis | |
| This needs to be rerun in order to reanalyze. | Sequencing data can be reanalyzed. |
| Need for Specific Personnel and Infrastructure | |
| Specific infrastructure and personnel are not required for microarray. | Specific infrastructure and personnel required by RNA sequencing. |
| Technical Issues | |
| Microarray technique has technical issues such as cross-hybridization, nonspecific hybridization, limited detection rate of individual probes, etc. | RNA seq technique avoids technical issues such as cross-hybridization, nonspecific hybridization, limited detection rate of individual probes, etc. |
| Biases | |
| This is a biased method since it depends on hybridization. | Bias is low compared to microarray. |
Summary – Microarray vs RNA Sequencing
Microarray and RNA sequencing methods are high throughput platforms developed for transcriptome profiling. Both methods produce results which are highly correlated to gene expression profiles. However, RNA sequencing has advantages over microarray for gene expression analysis. RNA sequencing is a more sensitive method for the detection of low abundance transcripts than microarray. RNA sequencing also enables the differentiation between isoforms and identification of gene variants. However, microarray is the common choice of most researchers since RNA sequencing is a new and expensive technique with data storing challenges and complex data analysis.
摘要–芯片与RNA测序
芯片和RNA测序方法是开发用于转录组分析的高通量平台。两种方法均产生与基因表达谱高度相关的结果。但是,RNA测序相对于基因表达分析而言,比微阵列具有优势。与微阵列相比,RNA测序是检测低丰度转录本的更灵敏方法。RNA测序还可以区分同工型和鉴定基因变体。然而,微阵列是大多数研究人员的常见选择,因为RNA测序是一种新的昂贵的技术,存在数据存储难题和复杂的数据分析。
References:
1.Wang, Zhong, Mark Gerstein, and Michael Snyder. “RNA-Seq: a revolutionary tool for transcriptomics.” Nature reviews. Genetics. U.S. National Library of Medicine, Jan. 2009. Web. 14 Mar. 2017
2.Rogler, Charles E., Tatyana Tchaikovskaya, Raquel Norel, Aldo Massimi, Christopher Plescia, Eugeny Rubashevsky, Paul Siebert, and Leslie E. Rogler. “RNA expression microarrays (REMs), a high-throughput method to measure differences in gene expression in diverse biological samples.” Nucleic Acids Research. Oxford University Press, 01 Jan. 2004. Web. 15 Mar. 2017
3.Zhao, Shanrong, Wai-Ping Fung-Leung, Anton Bittner, Karen Ngo, and Xuejun Liu. “Comparison of RNA-Seq and Microarray in Transcriptome Profiling of Activated T Cells.” PLOS ONE. Public Library of Science, Jan. 2014. Web. 15 Mar. 2017
Image Courtesy:
1. “Journal.pcbi.1004393.g002” By Malachi Griffith, Jason R. Walker, Nicholas C. Spies, Benjamin J. Ainscough, Obi L. Griffith – (CC BY 2.5) via Commons Wikimedia
2. “Microarray” By Bill Branson (Photographer) – National Cancer Institute (Public Domain) via Commons Wikimedia
最后
以上就是英勇香菇最近收集整理的关于关于微阵列芯片和RNA-seq的比较关于微阵列芯片和RNA-seq的比较Difference Between Microarray and RNA Sequencing的全部内容,更多相关关于微阵列芯片和RNA-seq的比较关于微阵列芯片和RNA-seq的比较Difference内容请搜索靠谱客的其他文章。







发表评论 取消回复