需注意的几点:
- 在无循环计算时:
令测试集为P(m×d),训练集为C(n×d),m为测试数据数据量,n为训练数据数据量,d是维度。
实际上最后距离公式变为如下形式
![]()
这里实现的依然是广播,所以只要P的形状为 m×1,C的形状为 1×n即可
- 三种循环方式体现的是运算速度的不同,结果是一致的
- k = 1时,training data自身的匹自身为100%
直接执行的代码(注释掉测试部分)
KNN中,训练即存储
import pickle
import os
import numpy as np
import matplotlib.pyplot as plt
from cs231n.data_utils import load_CIFAR10
from cs231n.classifiers import KNearestNeighbor
# This is a bit of magic to make matplotlib figures appear inline in the notebook
# rather than in a new window.
plt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
# Some more magic so that the notebook will reload external python modules;
# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython
# Load the raw CIFAR-10 data.
cifar10_dir = 'F:pycharmFileKNNcifar-10-batches-py'
X_train, y_train, X_test, y_test = load_CIFAR10(cifar10_dir)
# As a sanity check, we print out the size of the training and test data.
# print('Training data shape: ', X_train.shape)
# print('Training labels shape: ', y_train.shape)
# print('Test data shape: ', X_test.shape)
# print('Test labels shape: ', y_test.shape)
# Visualize some examples from the dataset.
# We show a few examples of training images from each class.
# classes = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']
# num_classes = len(classes)
# samples_per_class = 7
# for y, cls in enumerate(classes):
# idxs = np.flatnonzero(y_train == y) # 返回非零元素
# idxs = np.random.choice(idxs, samples_per_class, replace=False) # 随机不放回地抽出7个
# for i, idx in enumerate(idxs):
# plt_idx = i * num_classes + y + 1
# plt.subplot(samples_per_class, num_classes, plt_idx)
# plt.imshow(X_train[idx].astype('uint8'))
# plt.axis('off')
# if i == 0:
# plt.title(cls)
# # 查看样本
# plt.show()
# 取出子集, 用于train和test
# Subsample the data for more efficient code execution in this exercise
num_training = 5000
mask = range(num_training)
X_train = X_train[mask]
y_train = y_train[mask]
num_test = 500
mask = range(num_test)
X_test = X_test[mask]
y_test = y_test[mask]
# 将图像转置成二维
X_train = np.reshape(X_train, (X_train.shape[0], -1))
X_test = np.reshape(X_test, (X_test.shape[0], -1))
# print(X_train.shape, X_test.shape)
# KNN只将数据进行简单的存储
classifier = KNearestNeighbor()
#classifier.train(X_train, y_train)
# L2实现
#dists = classifier.compute_distances_two_loops(X_test)
# print(dists)
# print(dists.shape)
# 可视化training data和testing data的距离
# 深色表示距离小
# plt.imshow(dists, interpolation='none')
# plt.show()
# 开始test
# y_test_pred = classifier.predict_labels(dists, k=5)
# 打印正确率
# num_correct = np.sum(y_test_pred == y_test)
# accuracy = float(num_correct) / num_test
# print('Got %d / %d correct ==> accuracy: %f' %(num_correct, num_test, accuracy))
# 一层循环
# dists_one = classifier.compute_distances_one_loop(X_test)
# difference = np.linalg.norm(dists - dists_one, ord='fro')
# print('Difference was: %f' % (difference, ))
# if difference < 0.001:
# print('Good! The distance matrices are the same')
# else:
# print('Uh-oh! The distance matrices are different')
# 无循环
# dists_two = classifier.compute_distances_no_loops(X_test)
#
# difference = np.linalg.norm(dists - dists_one, ord='fro')
# print('Difference was: %f' % (difference, ))
# if difference < 0.001:
# print('Good! The distance matrices are the same')
# else:
# print('Uh-oh! The distance matrices are different')
# Let's compare how fast the implementations are
# def time_function(f, *args):
# """
# Call a function f with args and return the time (in seconds) that it took to execute.
# """
# import time
# tic = time.time()
# f(*args)
# toc = time.time()
# return toc - tic
# two_loop_time = time_function(classifier.compute_distances_two_loops, X_test)
# print('Two loop version took %f seconds' % two_loop_time)
#
# one_loop_time = time_function(classifier.compute_distances_one_loop, X_test)
# print('One loop version took %f seconds' % one_loop_time)
# no_loop_time = time_function(classifier.compute_distances_no_loops, X_test)
# print('No loop version took %f seconds' % no_loop_time)
通过交叉验证找到最好的k
# 交叉验证
num_folds = 5
k_choices = [1, 3, 5, 8, 10, 12, 15, 20, 50, 100]
X_train_folds = []
y_train_folds = []
X_train_folds = np.array_split(X_train, num_folds, axis=0)
y_train_folds = np.array_split(y_train, num_folds, axis=0)
# A dictionary holding the accuracies for different values of k that we find
# when running cross-validation. After running cross-validation,
# k_to_accuracies[k] should be a list of length num_folds giving the different
# accuracy values that we found when using that value of k.
k_to_accuracies = {}
for k in k_choices:
k_to_accuracies[k] = []
for val_idx in range(num_folds):
X_val = X_train_folds[val_idx]
y_val = y_train_folds[val_idx]
X_tra = X_train_folds[:val_idx] + X_train_folds[val_idx + 1:]
X_tra = np.reshape(X_tra, (X_train.shape[0] - X_val.shape[0], -1))
y_tra = y_train_folds[:val_idx] + y_train_folds[val_idx + 1:]
y_tra = np.reshape(y_tra, (X_train.shape[0] - X_val.shape[0],))
classifier.train(X_tra, y_tra)
y_val_pre = classifier.predict(X_val, k, 0)
right_arr = y_val_pre == y_val
accuracy = float(np.sum(right_arr)) / y_val.shape[0]
k_to_accuracies[k].append(accuracy)
# Print out the computed accuracies
for k in sorted(k_to_accuracies):
for accuracy in k_to_accuracies[k]:
print('k = %d, accuracy = %f' % (k, accuracy))
根据所得结果画出散点图,误差曲线
# plot the raw observations
print(k_choices)
print(k_to_accuracies)
for k in k_choices:
accuracies = k_to_accuracies[k]
plt.scatter([k] * len(accuracies), accuracies)
# 画出散点图和误差曲线
accuracies_mean = np.array([np.mean(v) for k,v in sorted(k_to_accuracies.items())])
accuracies_std = np.array([np.std(v) for k,v in sorted(k_to_accuracies.items())])
plt.errorbar(k_choices, accuracies_mean, yerr=accuracies_std)
plt.title('Cross-validation on k')
plt.xlabel('k')
plt.ylabel('Cross-validation accuracy')
plt.show()
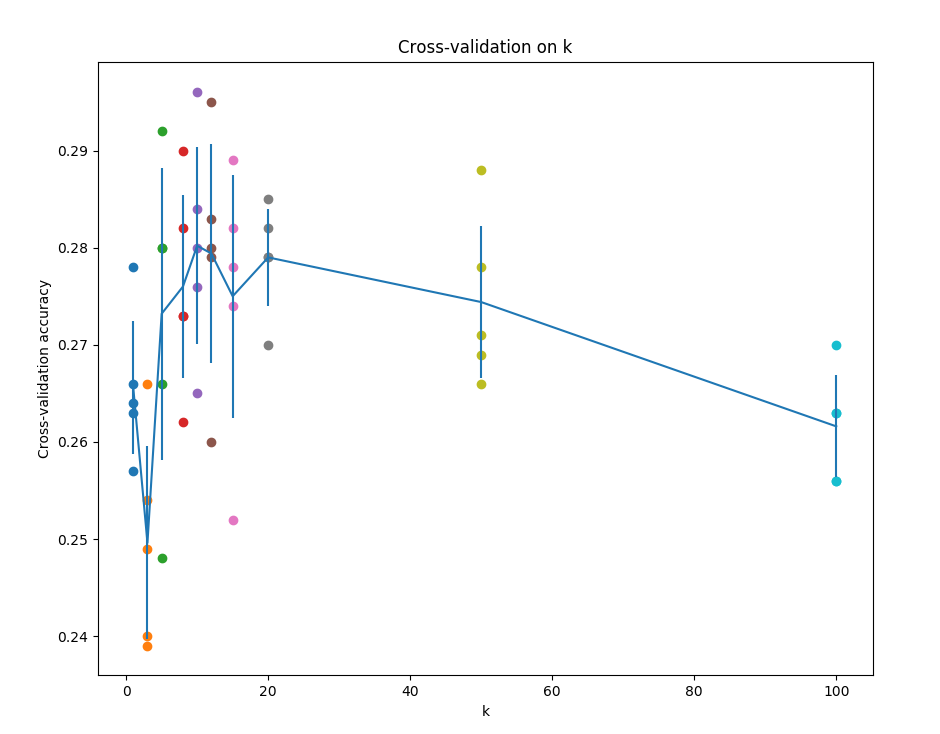
选择最好的k,此处当k=10时, 结果最好
best_k = 10
classifier = KNearestNeighbor()
classifier.train(X_train, y_train)
y_test_pred = classifier.predict(X_test, k=best_k)
# Compute and display the accuracy
num_correct = np.sum(y_test_pred == y_test)
accuracy = float(num_correct) / num_test
print('Got %d / %d correct => accuracy: %f' % (num_correct, num_test, accuracy))
修改原文件中的部分代码:
nearest_neighbor.py
import numpy as np
class KNearestNeighbor(object):
""" a kNN classifier with L2 distance """
def __init__(self):
pass
def train(self, X, y):
"""
Train the classifier. For k-nearest neighbors this is just
memorizing the training data.
Inputs:
- X: A numpy array of shape (num_train, D) containing the training data
consisting of num_train samples each of dimension D.
- y: A numpy array of shape (N,) containing the training labels, where
y[i] is the label for X[i].
"""
self.X_train = X
self.y_train = y
def predict(self, X, k=1, num_loops=0):
"""
Predict labels for test data using this classifier.
Inputs:
- X: A numpy array of shape (num_test, D) containing test data consisting
of num_test samples each of dimension D.
- k: The number of nearest neighbors that vote for the predicted labels.
- num_loops: Determines which implementation to use to compute distances
between training points and testing points.
Returns:
- y: A numpy array of shape (num_test,) containing predicted labels for the
test data, where y[i] is the predicted label for the test point X[i].
"""
if num_loops == 0:
dists = self.compute_distances_no_loops(X)
elif num_loops == 1:
dists = self.compute_distances_one_loop(X)
elif num_loops == 2:
dists = self.compute_distances_two_loops(X)
else:
raise ValueError('Invalid value %d for num_loops' % num_loops)
return self.predict_labels(dists, k=k)
def compute_distances_two_loops(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using a nested loop over both the training data and the
test data.
Inputs:
- X: A numpy array of shape (num_test, D) containing test data.
Returns:
- dists: A numpy array of shape (num_test, num_train) where dists[i, j]
is the Euclidean distance between the ith test point and the jth training
point.
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
for i in range(num_test):
for j in range(num_train):
#####################################################################
# TODO: #
# Compute the l2 distance between the ith test point and the jth #
# training point, and store the result in dists[i, j]. You should #
# not use a loop over dimension. #
#####################################################################
dists[i, j] = np.sqrt(np.sum(np.square(self.X_train[j] - X[i])))
#####################################################################
# END OF YOUR CODE #
#####################################################################
return dists
def compute_distances_one_loop(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using a single loop over the test data.
Input / Output: Same as compute_distances_two_loops
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
for i in range(num_test):
#######################################################################
# TODO: #
# Compute the l2 distance between the ith test point and all training #
# points, and store the result in dists[i, :]. #
#######################################################################
dists[i,:] = np.sqrt(np.sum(np.square(self.X_train - X[i,:]),axis=1))
#######################################################################
# END OF YOUR CODE #
#######################################################################
return dists
def compute_distances_no_loops(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using no explicit loops.
Input / Output: Same as compute_distances_two_loops
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
#########################################################################
# TODO: #
# Compute the l2 distance between all test points and all training #
# points without using any explicit loops, and store the result in #
# dists. #
# #
# You should implement this function using only basic array operations; #
# in particular you should not use functions from scipy. #
# #
# HINT: Try to formulate the l2 distance using matrix multiplication #
# and two broadcast sums. #
#########################################################################
dists = np.multiply(np.dot(X, self.X_train.T), -2)
sq1 = np.sum(np.square(X), axis=1, keepdims=True)
# keepdims = True, 使计算后依然为维度是测试样本数量的列向量、
sq2 = np.sum(np.square(self.X_train),axis=1)
dists = np.add(dists,sq1)
dists = np.add(dists,sq2)
dists = np.sqrt(dists)
#########################################################################
# END OF YOUR CODE #
#########################################################################
return dists
def predict_labels(self, dists, k=1):
"""
Given a matrix of distances between test points and training points,
predict a label for each test point.
Inputs:
- dists: A numpy array of shape (num_test, num_train) where dists[i, j]
gives the distance betwen the ith test point and the jth training point.
Returns:
- y: A numpy array of shape (num_test,) containing predicted labels for the
test data, where y[i] is the predicted label for the test point X[i].
"""
num_test = dists.shape[0]
y_pred = np.zeros(num_test)
for i in range(num_test):
# A list of length k storing the labels of the k nearest neighbors to
# the ith test point.
closest_y = self.y_train[np.argsort(dists[i])[:k]] #将矩阵按照axis排序,并返回排序后的下标
y_pred[i] = np.argmax(np.bincount(closest_y))
#########################################################################
# TODO: #
# Use the distance matrix to find the k nearest neighbors of the ith #
# testing point, and use self.y_train to find the labels of these #
# neighbors. Store these labels in closest_y. #
# Hint: Look up the function numpy.argsort. #
#########################################################################
pass
#########################################################################
# TODO: #
# Now that you have found the labels of the k nearest neighbors, you #
# need to find the most common label in the list closest_y of labels. #
# Store this label in y_pred[i]. Break ties by choosing the smaller #
# label. #
#########################################################################
pass
#########################################################################
# END OF YOUR CODE #
#########################################################################
return y_pred
最终结果:
![]()
KNN在物体识别,数据量有限的情况下,比较弱
最后
以上就是怕黑猎豹最近收集整理的关于cs231n assignment1 KNN的全部内容,更多相关cs231n内容请搜索靠谱客的其他文章。
本图文内容来源于网友提供,作为学习参考使用,或来自网络收集整理,版权属于原作者所有。








发表评论 取消回复