TRIM14 negatively affects proliferation/survival of NSCLC cell lines in vitro
We previously identified a prognostic multigene expression signature for early-stage NSCLC patients and hypothesized that these genes may function in the development and/or progression of NSCLCFig. 1a). Reduction in the representation of 20 shRNAs corresponding to genes essential for cell viability (e.g. CDK11B, RPS14, etc.) showed that the screen was functional (Supplemental Figure 1). Interestingly, downregulation of TRIM14 expression enhanced cell proliferation/survival in both H358 and H460 as represented by hairpin enrichment over time (Fig. 1a).
Figure 1: TRIM14 negatively affects proliferation/survival of NSCLC cell lines in vitro.

(a) Schematic diagram illustrates the experimental design of the pooled RNAi screen to identify genes affecting cell proliferation/survival in H460 and H358. Deep sequencing was used to quantify TRIM14 shRNA representation in H460 and H358 cell populations at multiple time points for two independent shRNAs. (b) Profiling TRIM14 protein expression in a panel of human NSCLC cell lines of different histological subtypes via Western blot analyses. (c) Whole-cell extracts from cell lines stably expressing either full-length TRIM14 or shRNAs against TRIM14 were subjected to Western blot analysis with anti-TRIM14 antibody and compared with isogenic shGFP or empty vector (EV) controls. β-actin served as a loading control. (d) TRIM14 downregulation and overexpression in cells (red) affects cell viability as measured by MTS assays. (f) Soft agar was used to assess the ability to form colonies in isogenic cell lines. Results shown represent three biological replicates. (Two-tailed student’s t-test, ****p
To validate the functional role of TRIM14 as a putative tumor suppressor gene (TSG) in lung cancer, we first screened TRIM14 protein expression using Western blot analysis in twelve different NSCLC cell lines and an immortalized but non-tumorigenic human bronchial epithelial cell line (HBE135). TRIM14 protein levels varied substantially among NSCLC cell lines with six out of twelve lines having lower expression levels compared to HBE135 cells (Fig. 1b).
We next established a panel of NSCLC isogenic cell lines that stably expressed the TRIM14 open reading frame (ORF) or shRNAs against TRIM14. H1650, H520, H157 and H358 cell lines, which endogenously express TRIM14, were transduced with two independent shRNAs (shTRIM14.A and shTRIM14.B) or a vector control (shGFP) and downregulation efficiency assessed by quantitative RT-PCR and Western blot analysis (Fig. 1c and Supplemental Figure 2). Both shTRIM14.A and shTRIM14.B mediated a clear decrease in TRIM14 protein levels relative to the vector control. We further overexpressed TRIM14 ORF constructs in H1395 and H3255 cell lines with nearly undetectable endogenous TRIM14 protein levels (Fig. 1c). Stable integration of shRNA or overexpression constructs did not alter the morphology of the cells, which remained epithelial in appearance.
Using MTS assays, we confirmed a significant increase in cell proliferation of H1650, H520 and H157 upon TRIM14 downregulation, consistent with our initial observation in H460 and H358 cells in the shRNA screen (Fig. 1d; p
Loss of TRIM14 contributes to tumorigenicity of NSCLC cell lines in vivo
We injected isogenic cell lines into subcutaneous tissue of scid mice to assess tumor growth rates. Overexpression of TRIM14 significantly attenuated tumor growth in H3255 xenograft models (Fig. 2a; p = 0.0021). Tumors formed by H3255-TRIM14 cells exhibited significantly reduced tumor weights than tumors formed by the empty vector controls (EV) at necropsy (Fig. 2b; p = 0.0314). Conversely, tumors formed by TRIM14-deficient cells (H1650 and H157) exhibited increased tumor growth rates and greater tumor weights than control tumors (Fig. 2c–e).
Figure 2: Loss of TRIM14 contributes to NSCLC tumorigenicity in mice.

(a) NSCLC cells were injected into the left flanks of 6–8 week-old SCID mice (n = 7–10 animals per group). Exogenous TRIM14 expression in H3255 significantly suppressed tumor growth in H3255-bearing mice. (b) Final tumor weights were measured for each H3255 tumor-bearing mouse at necropsy (EV = empty vector control). (c–d) Downregulation of TRIM14 in H157 and H1650 cells (red and blue) significantly increased tumor growth in mice. (e) Final tumor weights were measured for each H1650 tumor-bearing mouse at endpoint. (Two-way mixed ANOVA analyses for tumor growth rates and two-tailed student’s t-test for final weight measurements, ****p
Immunohistochemical (IHC) detection of cell proliferation marker Ki67 in H1650 xenograft tumor sections did not show a significant difference between control and experimental groups (Fig. 3b). However, staining using apoptotic marker cleaved caspase-3 demonstrated a significant albeit modest decrease in the number of cells with active caspase-3 in H1650-shTRIM14 xenograft tumors compared to controls (Fig. 3a,b; p = 0.0064). Moreover, Western blot analyses on tumor lysates confirmed sustained silencing of TRIM14 protein levels in H1650 during tumor progression, which coincided with reduced protein expression of apoptosis-inducing factor (AIF), another caspase-independent apoptotic pathway (Fig. 3c). Given that we observed apoptotic changes rather than expression of Ki67 or proliferating cell nuclear antigen (PCNA), we speculate that TRIM14 expression mediates apoptotic and cell death signaling. Together these results suggest that TRIM14 behaves as a TSG in NSCLC xenografts.
Figure 3: Reduced tumor growth of TRIM14-deficient cells correlates with decreased apoptotic activity in vivo.

(a,b) Representative histologic sections of xenografts of H1650 tumors were immunostained with Ki67 and cleaved-caspase-3 (CC3) antibody and the number of positive cells were quantified for 10 fields at high power (n = 10 controls; n = 8 shTRIM14.B; scale bar = 300 μm). (c) Total extracts from H1650 and H3255 xenograft tumors were subjected to Western blot analysis using the indicated antibodies and compared to isogenic controls. β-actin and GAPDH was used as loading controls. (Abbreviations: EV = Empty vector; Two-tailed student’s t-test; ****p
Association of TRIM14 expression with survival of NSCLC patients
We previously identified TRIM14 as a component of a prognostic multigene expression signature for early-stage NSCLC patientsTable 1). The four published microarray studies represented a total of 683 early-stage surgically resected NSCLC patients who had not received adjuvant chemotherapy or radiotherapy. Univariate survival analyses revealed that patients with high TRIM14 expression survived longer than patients with low expression in the BR.10 patient cohort (hazard ratio [HR] = 0.22, 95% CI = 0.09–0.56, p = 0.002) and the Director’s Challenge Consortium (DCC) cohort (HR = 0.61, 95% CI = 0.59–4.49, p = 0.026). TRIM14 expression did not correlate with survival in the University of Michigan study, which included only squamous carcinoma patients (HR = 1.17, 95% CI = 0.48–2.83, p = 0.733) (Table 1). Multivariate analysis revealed significant associations between TRIM14 expression and survival when adjusted for histological subtype, stage, age and sex in DCC and BR.10 cohorts. This data further supports the notion that TRIM14 down-regulation promotes NSCLC progression, which may lead to a poor clinical outcome.
Table 1 Clinical correlation of TRIM14 expression in NSCLC patients.
TRIM14 sensitizes NSCLC cells to anoxic-induced cell death
Given the biological implications of the in vivo results, we sought to understand the mechanism of TRIM14’s antitumor activity. We first analyzed the effect of TRIM14 downregulation on cell cycle progression by employing flow cytometry after propidium iodide staining. No significant effect on the distribution of G1, S or G2/M-phase cell populations was observed (Fig. 4a). Next, to determine whether TRIM14 may be inhibiting cell death pathways we assessed the effect of various apoptotic stimuli on TRIM14-deficient cells. While Staurosporine and Cisplatin treatment had no differential apoptotic effects on TRIM14-deficent cells compared to controls, apoptosis was significantly attenuated under anoxic conditions (<0.02% pO2 hypoxia) in TRIM14-deficient cells (H1650 and H358), suggesting that these cells were resistant to hypoxia-induced cell death (Fig. 4c,d; p
Figure 4: TRIM14 sensitizes NSCLC cells to anoxic-induced cell death and Type II interferon response.
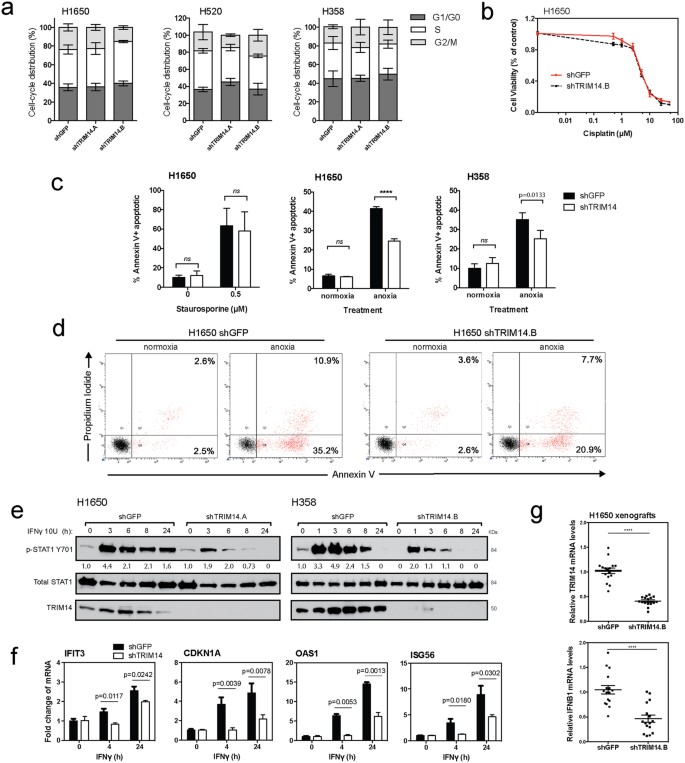
(a) Cell cycle progression of isogenic cell lines was assessed by flow cytometry after propidium iodide (PI) staining to determine the percent distribution of G1, S or G2/M-phase cell populations. (b) MTS assay was used to measure cell viability of H1650 cells treated with serial dilutions of cisplatin for 48 hours. (c) H1650 and H358 cells cultured for 48 hours with a protein kinase inhibitor, Staurosporine (0.5 μM), or under anoxic conditions were fixed and stained for Annexin-V and PI. Flow cytometry was subsequently used to quantitate the percentage of Annexin-V positive cells after treatment. (d) Representative flow cytometry analysis of H1650 cells cultured with or without anoxic conditions. (e) H1650 and H358 cells were treated with or without 10 U IFNγ for indicated times. Phosphorylation of STAT1 at tyrosine 701, total STAT1 and TRIM14 expression were analyzed by immunoblotting. (f) Quantitative RT-PCR using RNA extracted at 4 and 24 hours after IFNγ treatment showed increased transcript levels of ISG56, P21, IFIT1, and OAS1 as compared to untreated cells, but this effect was significantly suppressed in TRIM14-deficient cells. (g) Quantitative RT-PCR was used to show that mRNA levels for TRIM14 and IFNB1 were significantly reduced in H1650 xenograft tumors compared to controls (n = 8 tumors with two technical replicates each). Results shown represent more than three biological replicates. (Two-tailed student’s t-test; ****p
Loss of TRIM14 attenuates interferon response in NSCLC cells
Because previous studies have implicated the TRIMs as critical regulators in innate immune responses, we hypothesized that TRIM14 may exert antitumor cell effects via interferon signaling pathways in lung cancer cellsFig. 4e). Similar kinetics of STAT1 activation were observed in H358 cells when treated with IFNγ. These results suggest that TRIM14 is required for STAT1 activation following IFNγ stimulation. TRIM14 downregulation also significantly decreased transcription of well-characterized IFNγ-induced genes and STAT1 downstream targets in H1650 cells upon IFNγ treatment (Fig. 4f). In control cells, quantitative RT-PCR using RNA extracted at 4 and 24 hours after IFNγ treatment showed significantly increased levels of IFIT3, CDKN1A, OAS1 and ISG56 transcripts as compared to vehicle-treated cells. However, this enhancement was attenuated in the TRIM14-deficient cells indicating that TRIM14 is critical for STAT1-dependent gene expression.
Furthermore, recent studies have reported impaired type I IFN production in TRIM14−/− mice upon herpes simplex (HSV) infectionFig. 4g; p
Regulation of TRIM14 stability by proteasome degradation
To identify TRIM14 interacting proteins, we employed immunoprecipitation (IP) coupled with LC-MS/MS. HEK293T cells were transiently transfected with the full length TRIM14 ORF tagged at the C-terminus with 3xFLAG. FLAG-tagged TRIM14 complexes were isolated from total protein lysate by immunoprecipitation 48 hours after transfection and eluted proteins were digested with trypsin and characterized by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using a Protein Prophet p-value cutoff of > 0.95Table 2). To validate our MS shortlist, we performed co-immunoprecipitation assays to show that TRIM14 bound to AIF and HAX1 in HEK293T cells (Fig. 5a,b). Ectopically expressed FLAG-TRIM14 immunoprecipitated with endogenous p62 in both HEK293T and H1395 cells (Fig. 5c,d). We next identified enriched Gene Ontology (GO) functions in this interactome by performing David bioinformatics (Supplemental Table 5). Enrichment analysis revealed significantly enriched canonical pathways associated with: (1) protein ubiquitination, (2) programmed cell death, and (3) regulation of apoptosis.
Table 2 Top 70 proteins that co-immunoprecipitated with TRIM14 in HEK293T cells.
Figure 5: TRIM14 protein stability is regulated by ubiquitin-proteasome system.

(a) Whole-cell extracts from HEK293T cells transfected with either empty FLAG vector or vector ectopically expressing FLAG-TRIM14 were immunoprecipitated with anti-FLAG antibody, and immune complexes was blotted with indicated antibodies. The ectopically expressed TRIM14 bound with endogenous AIF and HAX1 in HEK293T cells. (b) Conversely, endogenous TRIM14 was immunoprecipitated using anti-AIF antibody in HEK293T cells. (c) Ectopically expressed FLAG-TRIM14 immunoprecipitated with endogenous p62 in HEK293T and (d) H1395 cells. (e) Mass spectrometry analysis of FLAG-TRIM14 complexes independently showed that TRIM14 interacted with a number of E3 ubiquitin ligases and proteasome activators in HEK293T cells. (f) HEK293T cells were transiently transfected with the indicated vectors and were treated with or without 10 μM MG132 for 4 hours demonstrating that ubiquitylation of FLAG-TRIM14 is elevated in the presence of MG132. (g) HEK293T cells were treated with either DMSO or 10 μM MG132 for 4 hours before the addition of 100 μg/ml of cycloheximide (CHX). Immunoblot analysis was performed on HEK293T cell lysates at the indicated times post-cycloheximide treatment to determine TRIM14 protein stability. (h) Immunoblot analysis was done on H520 cells treated with either vehicle (DMSO) or Bortezomib (10 nM) at indicated time points. Abbreviations: IgG = immunoglobulin G, IP = Immunoprecipitation and IB = Immunoblotting.
Given the abundance of E3 ubiquitin ligases and proteasome activators that consistently immunoprecipitated with TRIM14, we postulate that TRIM14 protein levels may be regulated by the proteasome (Fig. 5e). To examine whether TRIM14 is ubiquitylated in vivo, we ectopically expressed both HA-ubiquitin and FLAG-TRIM14 in HEK293T cells in the presence or absence of the proteasome inhibitor MG132. Addition of MG132 enhanced the levels of ubiquitin linked to FLAG-TRIM14 (Fig. 5f). We then examined the effect of proteasome inhibition on the stability of TRIM14. HEK293T cells were treated with either DMSO or MG132 and harvested at indicated time intervals after cycloheximide (CHX) treatment (Fig. 5G). The half-life of TRIM14 was ~6 hours in the absence of proteasome inhibition but was extended to >12 hours in the presence of MG132, indicating that the proteasome is required for rapid turnover of TRIM14 protein levels. Furthermore, treatment with a more potent proteasome inhibitor, Bortezomib, on H520 cells enhanced TRIM14 protein levels after 6 hours on treatment (Fig. 5h).
最后
以上就是正直草莓最近收集整理的关于signature=a6941c320414a50941f39ed49eae9b2f,TRIM14 is a Putative Tumor Suppressor and Regulator of In...的全部内容,更多相关signature=a6941c320414a50941f39ed49eae9b2f,TRIM14内容请搜索靠谱客的其他文章。








发表评论 取消回复